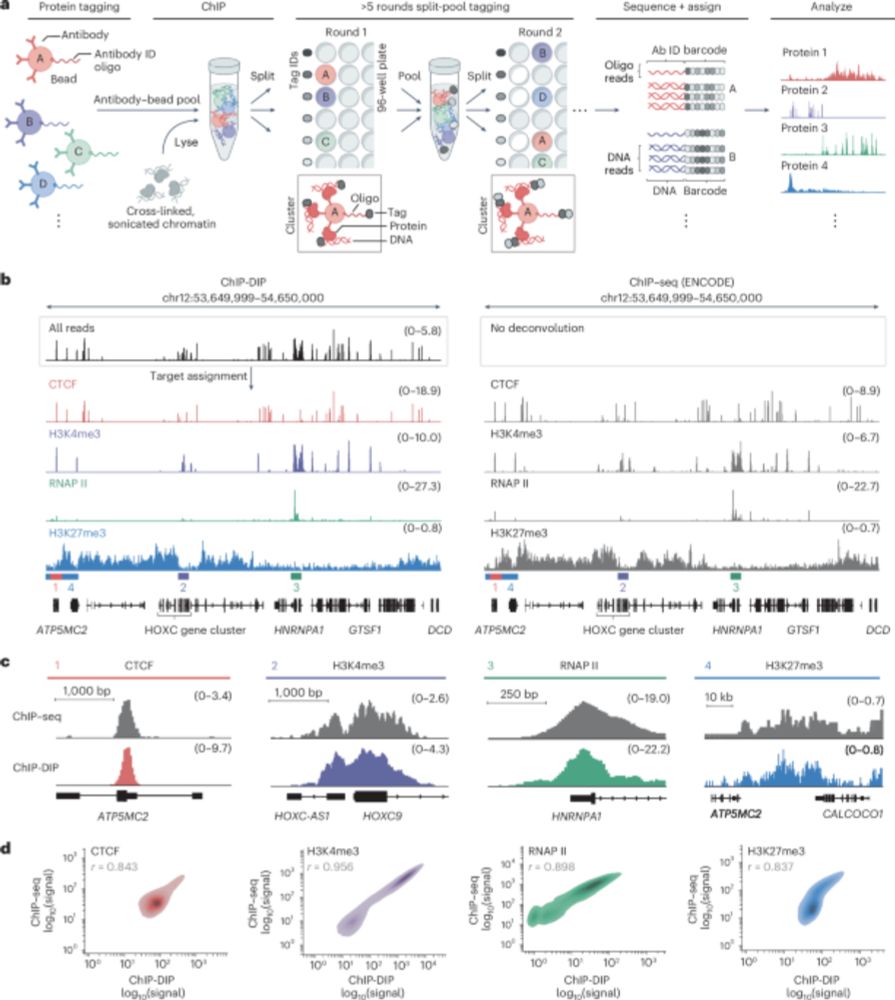
Epigenetics Update - MethAgingDB: a comprehensive DNA methylation database for aging biology go.nature.com/4ltmg2T
Haixin Wang and Shengquan Chen reporting in Scientific Data
#Epigenetics #DNAm #DNAme #Aging
---
Empower your research with high-res epigenetic insights at epigenometech.com
18.07.2025 13:02 — 👍 0 🔁 1 💬 0 📌 0
Graphical abstract of the paper.
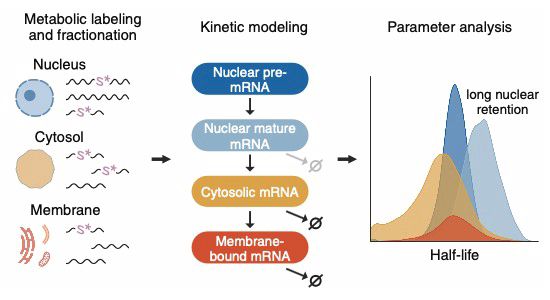
How long does an mRNA stay in the nucleus? How long does it stay in the cytoplasm? In an amazing collaboration with @landthalerm.bsky.social, we used metabolic labeling, cell fractionation and mathematical modeling to quantify mRNA flow through the cell. Finally out in MSB: doi.org/10.1038/s443...
21.11.2024 14:56 — 👍 217 🔁 87 💬 12 📌 10
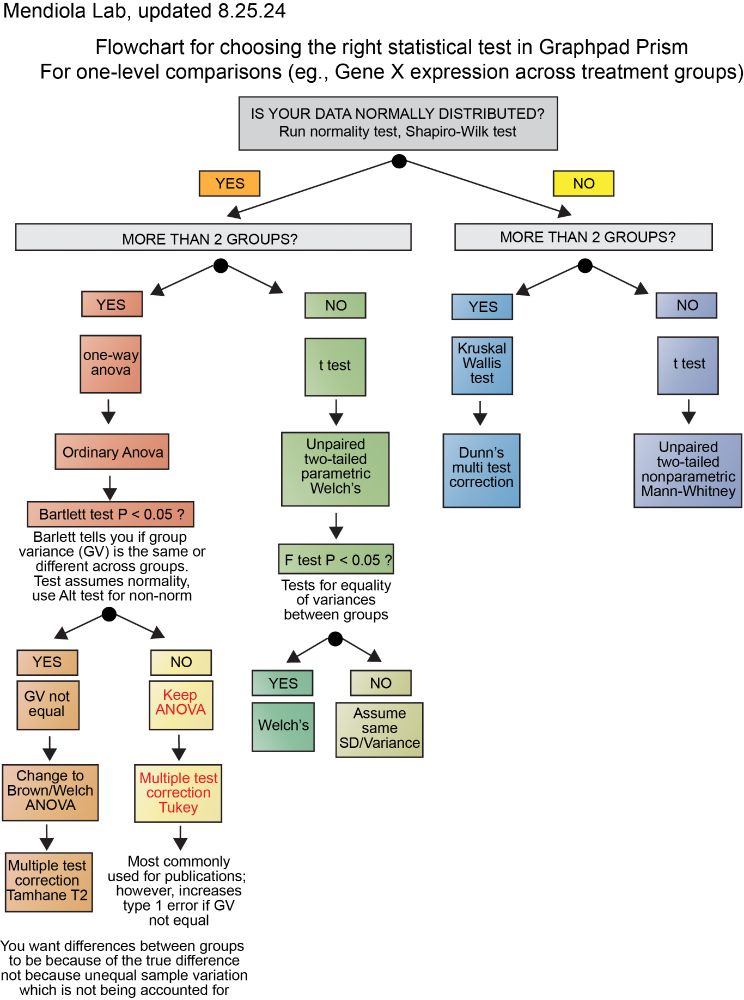
I created a simple flow chart to help my lab trainees choose the most appropriate stats test in Prism (*for single-factor comparisons only). Feel free to share with your group if you think it could be helpful first start.
20.11.2024 17:20 — 👍 258 🔁 89 💬 20 📌 3

Our recent study in Cell Metabolism provides compelling evidence that chromatin accessibility and transcription factor network remodeling in aging reflect the predictable degrading effects of a mechanism initially driving organismal maturation.
Link: doi.org/10.1016/j.cmet.2024.06.006
Thread 🧵👇1/9
18.11.2024 10:09 — 👍 97 🔁 25 💬 3 📌 3
A few years old, but worth a read (perhaps in particular to those who do ML in BIO or take biological databases for granted) 🧪🧬🖥️
"How Margaret Dayhoff Brought Modern Computing to Biology"
www.smithsonianmag.com/science-natu...
12.11.2023 13:26 — 👍 30 🔁 17 💬 1 📌 0
Leonid Mirny and I wrote this for all interested in chromosomes: "The chromosome folding problem and how cells solve it"
www.cell.com/action/showP...
14.11.2024 16:02 — 👍 67 🔁 41 💬 0 📌 2
I’ve just read this pre-print and it’s fantastic. A conserved element with no endogenous enhancer activity is nonetheless necessary and sufficient for extreme long range enhancer action. Recommend to anyone interested in gene regulation
13.11.2024 21:15 — 👍 29 🔁 5 💬 1 📌 1
Created one! Let me know if you find anyone that is missing.
go.bsky.app/StHb8Ma
09.11.2024 02:49 — 👍 11 🔁 7 💬 14 📌 1
Hi folks on Bluesky! I'm a postdoc at Harvard Medical School, studying epigenomics underlying aging. Specifically, I am trying to understand the spatiotemporal dynamics of partial reprogramming by three of four Yamanaka factors (OSK). Happy to be here!
13.11.2024 20:41 — 👍 6 🔁 0 💬 0 📌 0
ok, I tried to make a starter pack on genome stability/DNA replication/DNA repair etc. Sorry if I couldn't find someone, please nominate yourself and your colleagues in comments or DMs, looking forward to connecting with everyone!
go.bsky.app/9jdDNxG
12.11.2024 01:11 — 👍 124 🔁 65 💬 44 📌 4
Hi BlueSky! It's great to be here. Here's a handy Starter Pack of DNA repair & genome stability groups that I found so far go.bsky.app/NvFWKvL
13.11.2024 09:03 — 👍 54 🔁 22 💬 10 📌 4
Hi Bluesky! I made a 3D chromatin starter pack.
Let me know if you would like to be added.
go.bsky.app/6tTQdqQ
13.11.2024 17:55 — 👍 84 🔁 42 💬 41 📌 0
We are the Ting Wu Lab @ HMS. Our laboratory studies how chromosome behavior and positioning influence genome function, with implications for gene regulation, genome stability, and disease.
Managed by WuLab members
https://www.transvection.org/home
🧬 The Genome • Unrestricted 🧪
Epigenome Technologies, pioneering in epigenetic and single-cell analysis, offers advanced assay kits and optimization services.
Director of bioinformatics at AstraZeneca. subscribe to my youtube channel @chatomics. On my way to helping 1 million people learn bioinformatics. Educator, Biotech, single cell. Also talks about leadership.
tommytang.bio.link
Senior Scientist, Department of Biology, Emory University, Atlanta, GA.
Molecular Biologist. RNA Scientist. Yeast Geneticist 🧬. British🇬🇧. Runner 🏃🏻…. Opinions are my own.
Interests: RNA Decay, RNA Processing, RNA & Disease.
Cell biology lab at Newcastle University, UK, molecular mechanisms of nutrient sensing and autophagy, ageing and age-related disease 🇺🇦
Exploring brain aging with data science and 'omics.
https://ddfishbean.github.io/
PhD Candidate at Tae-Young Yoon Lab,
Seoul National University, South Korea
# Single-molecule Biophysics, Drug Discovery, Biomolecular Condensate, Transcription factor, Cancer and Aging-related Diseases
Neural crest biologist interested in broad and deep questions about clockworks of nature.
Molecular biology lab based at the Department of Biology, University of Oxford. Researching insect epigenetics and epigenetic inheritance. Actively recruiting!
Postdoc with @Giacomo_Cavalli, former PhD student in the @marstonlab.bsky.social. HFSP Fellow, EMBO Fellow. 3D genome enthusiast!
Staff Scientist @ Ludwig Princeton Branch
Cancer Immunotherapy & Harder Family Chair Cancer Research #FoxLab @ChilesResearch /CEO @UbiVac / @OHSUmmi.bsky.social /exPres&Ambassador @SITCancer.bsky.social /ex #NCI /ex #Detroit /M GO BLUE/ WIC & JEDI/ @detroitlions.bsky.social /❤️Pinot/Tweets my own
Analytical biochemist attempting molecular cartography across the tree of life, powered by MS-based proteomics. Opinions expressed are solely my own. he/him
benjaminneely.com
Charleston, SC, USA
Scientist, leader and carer. System biology, Epigenetics, Pluripotency, Immunology but mostly eXIST! Wondering why men and women are so different. Marie-Curie, EMBO, HFSP alumnus. Tenure Associate Prof/Group Leader. Editor ScienceAdvances. Mamma di Sofia!
GenX Physical Therapist, wife, NYUmom, taco lover, golf, student of life. Creator of online Pilates inspired workouts for bodies that have “been places.” www.bewellstaywell.net. Movement & cultural retreats in #PuertoVallarta, #golffitness #PilatesSky
Dad x 2, husband, son, brother, computational biologist, genome scientist, associate professor at la jolla institute for immunology (LJI) and UCSD https://www.lji.org/labs/ay/
Epigenetics, transposable elements, plant biology.
Postdoc @ Gregor Mendel Institute (Vienna, Austria).
@gmivienna.bsky.social
When I grow up, I want to be a scientist. Postoc in @grothlab.bsky.social. Fond of chromatin biology, DNA replication, space, pandas, and more. PhD from @lab-hat.bsky.social.