Ligand Binding Free Energy Landscapes at the Tubulin Colchicine Site from Coarse-Grained Metadynamics https://www.biorxiv.org/content/10.64898/2026.02.24.707696v1
26.02.2026 02:47 —
👍 2
🔁 2
💬 0
📌 0
Our Story about ADAM10 modulation by PS lipids found its home in Advanced Science !
Many thanks to all co-authors for an amazing collaboration.
24.02.2026 19:14 —
👍 7
🔁 3
💬 0
📌 0
YouTube video by Marieke
Bentopy tutorial walkthrough
I just uploaded a video walkthrough of the tutorial I recorded this morning.
youtu.be/C4LYZokS_t4
13.02.2026 16:41 —
👍 6
🔁 3
💬 0
📌 0
Big thanks to all the coauthors, especially @ma3ke.bsky.social for driving this paper forward. Really nice to have this work finally out there, and especially cool that it's part of the special issue honoring the work of David Goodsell. A huge inspiration for whole-cell modeling!
13.02.2026 15:37 —
👍 4
🔁 0
💬 0
📌 0

Bentopy: from simple packing to building cellular models – Martini Force Field Initiative
Beyond the minimal cell, we also packed the multi-compartment architecture of a mitochondrion and built an all-atom SARS-CoV-2 virion in a respiratory aerosol (based on work from the Amaro lab). Want to try it yourself? We wrote a tutorial on the Martini website, give it a go!
13.02.2026 15:37 —
👍 2
🔁 0
💬 1
📌 0
Building realistic models of cells means fitting proteins, RNA, and metabolites at experimental concentrations inside complex cellular architectures. Here's a movie showing how we pack the cytoplasmic space of the JCVI-Syn3A cell:
13.02.2026 15:37 —
👍 6
🔁 4
💬 2
📌 1

Cutaway and close-up views of a Martini coarse-grained whole-cell model of JCVI-syn3A, showing the densely packed cytoplasm with proteins, RNA, metabolites, and chromosome inside a lipid membrane with embedded membrane proteins
Our paper on [Bentopy](doi.org/10.1002/pro....) is out in Protein Science! We developed Bentopy to make assembling large-scale MD models more accessible, building on what we learned from trying to simulate whole-cell models. Here's our updated Martini JCVI-syn3A cell model👇
13.02.2026 15:37 —
👍 32
🔁 17
💬 1
📌 0
So pleased this is finally out! Wonderful work by the incredibly talented @ma3ke.bsky.social and @janstevens.bsky.social to generate densely packed systems, even with complex topologies
Go to the paper for cool science, stay for the mesmerizing figures 🤩
Read more about it in this thread 👇
13.02.2026 15:22 —
👍 7
🔁 3
💬 0
📌 0

Model of mitochondrial compartments. The assembly of the mitochondrial model based on an experimentally informed membrane structure (white), from an empty structure (left) into a mask representation of the IMS (yellow) and matrix (pink) compartments represented by 3 nm voxels. Structures are packed into their assigned compartments based on the mask. In the last section and the magnified inset, structures are colored by kind: Proteins (green), RNA (dark blue), metabolites (pale blue).
Our paper about Bentopy is now published in Protein Science!
Bentopy makes assembling large-scale MD models accessible and fast.
doi.org/10.1002/pro....
@janstevens.bsky.social, @cg-martini.bsky.social
13.02.2026 15:07 —
👍 13
🔁 6
💬 1
📌 1
It was great to be a small part of this work with the incredibly talented PhD student Andrea!!
If you want to find out how you can save heaps of time while still maintaining accurate results with CG metadynamics, look no further!! Read the article here 👇
12.01.2026 10:11 —
👍 8
🔁 1
💬 0
📌 0
Instead of a tree, I can offer a snowman! ☃️
Built using bentopy, based on David Naranjo's idea.
The input file can be found here: gist.github.com/ma3ke/723f9c....
24.12.2025 16:35 —
👍 7
🔁 3
💬 1
📌 0
Opportunity: postdoctoral fellowships at the center for Quantitative Cell Biology (QCB), Univ. of Illinois, in close collaboration with our group. Your ticket to become an expert in whole-cell Martini simulations !
qcb.illinois.edu/postdoctoral...
22.12.2025 10:27 —
👍 7
🔁 3
💬 0
📌 0
My little outlook on how our lungs 🫁 are like the sea 🌊 is finally out in Central Science. Thanks to the support from incredible mentors @rommieamaro.bsky.social and @kprather.bsky.social !!
11.12.2025 12:18 —
👍 12
🔁 8
💬 0
📌 0

A new review explores one of the themes of my book Air-Borne: our lungs are like the ocean. Both send life into the atmosphere. pubs.acs.org/doi/full/10....
07.12.2025 21:26 —
👍 80
🔁 22
💬 2
📌 3
Congrats Pavol! That poster looks incredible, amazing design 🎉
27.11.2025 14:08 —
👍 3
🔁 0
💬 0
📌 0
Call for Applications: Early Career Rescue Fellowship – Freiburg Institute for Advanced Studies
Are you a postdoc stuck in the US?! Is your research stalled or are you in any way affected by the whims of that unfettered madman? Interested in MD simulations of large biomolecules?!
Consider joining me at the University of Freiburg!
uni-freiburg.de/frias/call-f...
17.11.2025 08:51 —
👍 16
🔁 16
💬 0
📌 0

Beautiful picture looking out of the chemistry building at the University of Freiburg in Freiburg Germany. The old cathedral can be seen in the distance with a backdrop of a blue sky with clouds and distant foothills of the black forrest with fall colors showing through in the trees

picture of the Münster at dawn in the Altstadt of Freiburg, Germany, with a still dark sky in the background and moon over the Münster, in the foreground a well-lit flowers stand can be seen preparing for the daily morning market
I have a PhD position available in my lab at the University of Freiburg!! If you, or someone you know, might be looking for a position using modeling and simulations to uncover secrets in (glyco-)protein structure/function relationships please take a look at our website!
www.kearnslab.org
17.10.2025 20:21 —
👍 22
🔁 19
💬 0
📌 0
Inferring DNA kinkability from biased MD simulations https://www.biorxiv.org/content/10.1101/2025.09.08.674847v1
12.09.2025 21:47 —
👍 1
🔁 1
💬 0
📌 0

4️⃣ Featuring the fourth of our showcase projects
Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology
12.09.2025 06:21 —
👍 29
🔁 11
💬 1
📌 1
New preprint: www.biorxiv.org/content/10.1...
We used molecular dynamics simulations and solid-state NMR (@fmp-berlin.de) to look at calcium binding to the pore domain of the calcium-gated K+ channel MthK. @wojciechkopec.bsky.social @compbiophys.bsky.social
19.08.2025 07:42 —
👍 17
🔁 6
💬 1
📌 2
Weird lipids, weird effects on membranes ! Happy to see this story out 😄
If you like MD simulations, NMR spectroscopy and membranes this story will interest you 😊
11.08.2025 13:44 —
👍 12
🔁 3
💬 0
📌 0
Classification of containment hierarchy for point clouds in periodic space https://www.biorxiv.org/content/10.1101/2025.08.06.668936v1
10.08.2025 05:47 —
👍 2
🔁 2
💬 0
📌 0
Happy to share the latest work from the lab, led by @mudgal17.bsky.social, in collaboration with the Weis lab @ethzurich.bsky.social.
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
23.07.2025 11:40 —
👍 141
🔁 37
💬 5
📌 3
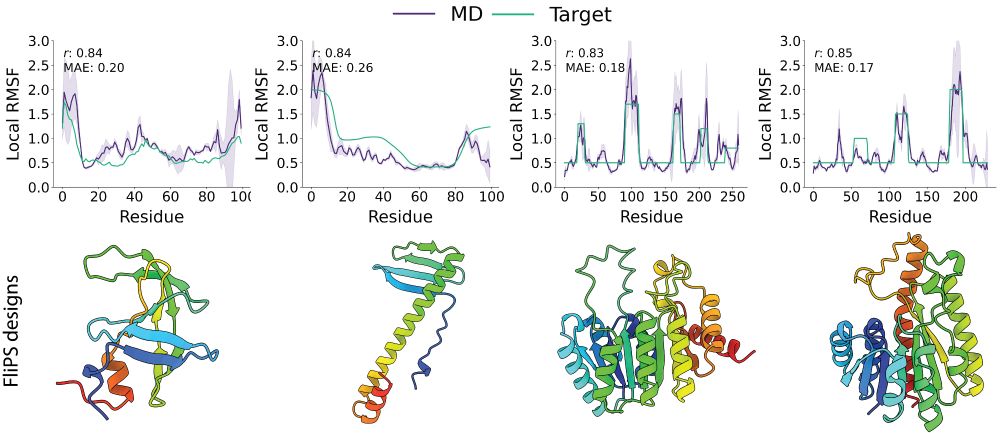
1/6 One of the key features of functional proteins is their inherent structural flexibility. In our recent work at #ICML, we introduce flexibility to protein structure design! More in a thread below.
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
17.07.2025 04:18 —
👍 22
🔁 8
💬 1
📌 3
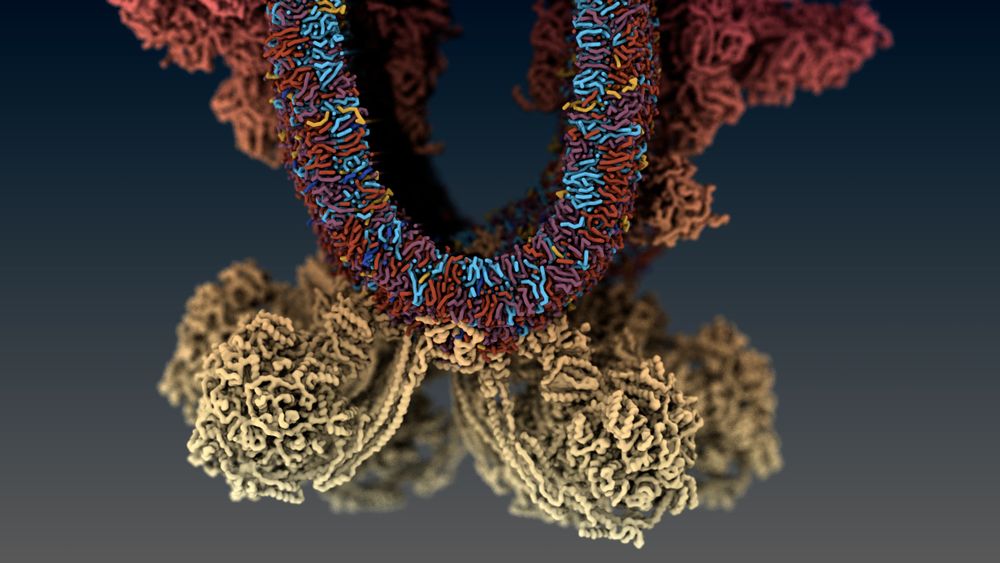
A stylized image of the molecular model of a mitochondrial crista, with proteins and lipids shown
What better way to use my first proper post than to share my first big piece of post-doctoral work with @cg-martini.bsky.social!
Here, we used integrative modelling to build and simulate a mitochondrial cristae.
Find the paper here (rdcu.be/eujAC) or see below for a quick overview 👇
01.07.2025 15:33 —
👍 49
🔁 14
💬 4
📌 0

















