We got a new preprint just published. Here, we redesigned double-stranded RNA binding domains to inhibit the RNA helicase DHX9 through a highly specific interface. A proof of principle for inhibitor design based on autoregulatory domains. www.biorxiv.org/content/10.6...
16.02.2026 06:17 —
👍 4
🔁 0
💬 0
📌 0

Peer Bork - personal thoughts | Ewan Birney
Personal thoughts on the passing of Peer Bork
I have been one of the many people across the world impacted by the sudden death of Peer Bork - a friend, colleague and remarkable scientist. I now have enough time this weekend to write some personal thoughts about Peer. www.linkedin.com/posts/ewan-b...
24.01.2026 15:26 —
👍 29
🔁 10
💬 2
📌 0
The story of how multiple RBPs cooperate to repress translation is now out in @cp-cellreports.bsky.social by @marcopayr.bsky.social and in a great collaboration with @hennig-lab.bsky.social!
www.cell.com/cell-reports...
07.11.2025 15:34 —
👍 42
🔁 15
💬 3
📌 0

Congrats to the Grünewald, Topf groups & et. al @cssbhamburg.bsky.social on their Nature paper! 📃 🍾
Trapping and resolving the elusive prefusion form of HSV gB - neutralizing HSV-1 & HSV-2 with a single nanobody is no small feat!🦙
Glad that our SPC facility have contributed to this work!@embl.org
08.09.2025 13:57 —
👍 5
🔁 3
💬 0
📌 0
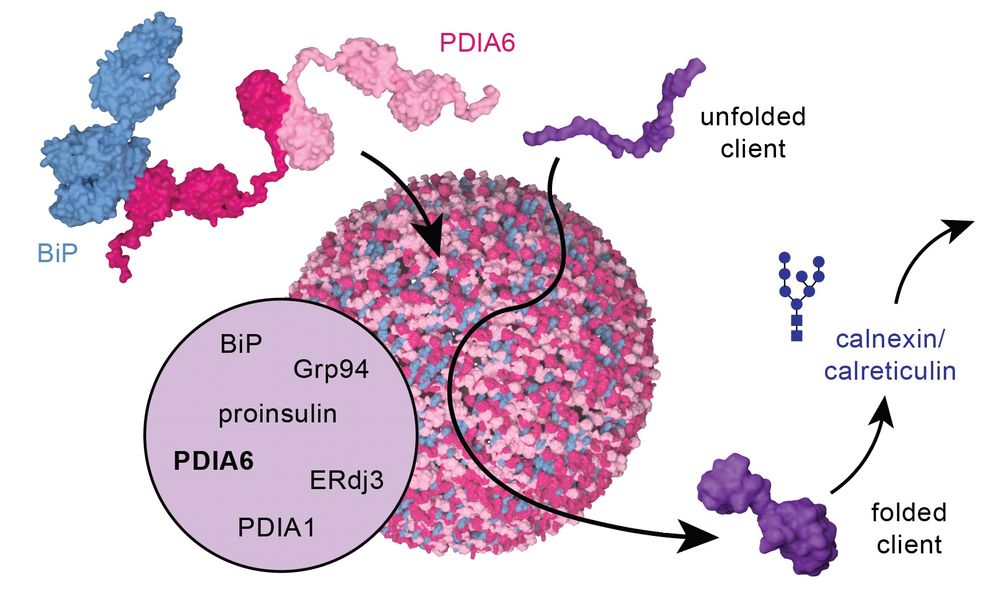
Happy to share my first publication, where we present our discovery of multi-chaperone condensates that orchestrate protein folding in the endoplasmic reticulum!
www.nature.com/articles/s41...
11.08.2025 15:07 —
👍 64
🔁 18
💬 5
📌 2

"Hey Bob, just dock the AF3 prediction into density and deposit"
07.08.2025 17:04 —
👍 28
🔁 3
💬 3
📌 0

With Instagram-ready spike ball pictures (pay attention to the sun beam! 😇).
29.07.2025 15:50 —
👍 0
🔁 0
💬 0
📌 0

Instagram-ready group picture from our lab hike
29.07.2025 15:48 —
👍 3
🔁 0
💬 1
📌 0
Indeed!
11.07.2025 15:00 —
👍 1
🔁 0
💬 0
📌 0

The most expensive bike stand in the world! 😢
11.07.2025 14:21 —
👍 9
🔁 1
💬 3
📌 0
Happy to see @embl.org highlight our work on protein design at Korbel group with @typaslab.bsky.social and @hennig-lab.bsky.social !
Working with everyone was a blast and helping to bring our software to production at DenovAI was quite the exciting experience.
26.06.2025 02:24 —
👍 11
🔁 2
💬 0
📌 0
Happy to announce the first paper from my PhD at Korbel group at @embl.org has finally been published:
embopress.org/doi/full/10.1038…
Collaborating with @typaslab.bsky.social, @hennig-lab.bsky.social and the EMBL PEPCF, we designed de novo inhibitors to a bacterial phage defense system 1/🧵
19.06.2025 08:10 —
👍 30
🔁 7
💬 2
📌 3
Check it out if you are interested in designing your own protein binders to any protein target. It works (see example in the article).
17.06.2025 14:02 —
👍 1
🔁 0
💬 0
📌 0
We did it in 8:43 h! A great team and leaders. So much fun. Thanks to everyone who has donated 😍! If you still want to donate it is possible until this Friday (20th of June): egeninsamling.brostcancerforbundet.se/fundraisers/...
16.06.2025 12:04 —
👍 1
🔁 0
💬 0
📌 0

After Vätternrundan

Medal show-off ;-)

Splits at different time checks

Optimal conditions :-)
Last Saturday I rode the Vätternrundan with the charity organization "I move for cancer" to raise money for breast cancer research. The goal was to finish the ride of 315 km around beautiful lake Vättern faster than 9 h.
16.06.2025 12:04 —
👍 4
🔁 1
💬 1
📌 0

Single-molecule multimodal timing of in vivo mRNA synthesis
mRNA synthesis requires extensive pre-mRNA maturation, the organisation of which remains unclear. Here, we directly sequence pre-mRNA without metabolic labelling or amplification to resolve transcript...
The timeline of co-transcriptional #mRNA processing and #modification may have to be revised: cleavage, termination, and #m6A deposition may occur earlier than most #splicing! Potential textbook changing study in bioRxiv by edueyras.bsky.social & Rippei Hayashi labs!
www.biorxiv.org/content/10.1...
29.04.2025 20:31 —
👍 55
🔁 21
💬 1
📌 1
“We've arranged a society based on science and technology, in which nobody understands anything about science and technology. And this combustible mixture of ignorance and power, sooner or later, is going to blow up in our faces.”
~ Carl Sagan
26.04.2025 13:51 —
👍 2055
🔁 661
💬 34
📌 70
Looks like the evidence for this weird chemical abundance in an exoplanet atmosphere has gotten stronger with new JWST data: www.nytimes.com/2025/04/16/s...
Is it really a sign of life? That’s still not clear; lots of analysis left to do. But it’ll be interesting to see how it all pans out! 🧪 🔭
17.04.2025 03:00 —
👍 1995
🔁 350
💬 42
📌 39
Ever wondered how several individual RBPs cooperate to repress translation?
Lead by @marcopayr.bsky.social and in great collaboration with @hennig-lab.bsky.social, we simultaneously tracked the binding of several proteins to single mRNA molecules in real-time!
www.biorxiv.org/content/10.1...
12.04.2025 11:29 —
👍 53
🔁 17
💬 0
📌 3
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
11.04.2025 08:35 —
👍 360
🔁 141
💬 12
📌 20
🧵 8/8 Combining different methodologies (smFRET, NMR, reporter gene assays) we developed a framework that can now be used to study similar processes in other biological systems! 🧬 #single-molecule #FRET #RNA #translation #Biophysics
11.04.2025 04:42 —
👍 1
🔁 0
💬 0
📌 0
🧵 7/8 Why this matters: Understanding these molecular mechanisms helps us grasp how cells regulate their genes through different complex assembly mechanisms
11.04.2025 04:41 —
👍 0
🔁 0
💬 0
📌 0






















