Electrostatic changes enabled the diversification of an exocyst subunit via protein complex escape - Nature Plants
The evolutionary diversification of an exocyst subunit was enabled by electrostatic shifts leading to its dissociation from the ancestral complex.
My main work as postdoc @plantophagy.bsky.social lab in @gmivienna.bsky.social is out in @natplants.nature.com 🌱🎉
We asked how can protein complexes diversify without compromising their function and explored this question using the plant #exocyst complex.
www.nature.com/articles/s41...
31.10.2025 13:57 — 👍 72 🔁 34 💬 5 📌 1
Congratulations Juan. Really amazing 🤩
01.11.2025 10:26 — 👍 1 🔁 0 💬 1 📌 0
Fair.
It is just easier for me to believe that someone tries to get away with something like this than someone actually getting away with it (for now at least). Mind boggling.
15.10.2025 21:35 — 👍 1 🔁 0 💬 1 📌 0
Whatever happened here cannot be called peer review imo. I don't know who did worse - the authors or the "reviewers". They even show the frickin map in a main figure.
Crazy that many excellent papers in Structure have to stand next to something like this.
15.10.2025 19:18 — 👍 6 🔁 0 💬 1 📌 0

Figure 1 from the preprint
Check out our preprint! With new molecular mechanisms, 140 subtomogram averages, and ~600 annotated cells under different conditions, we @embl.org were able to describe bacterial populations with in-cell #cryoET. And there’s a surprise at the end 🕵️
www.biorxiv.org/content/10.1...
#teamtomo
15.10.2025 06:26 — 👍 113 🔁 34 💬 8 📌 4
Thrilled to share that I’ll be joining @imbmainz.bsky.social in February 2026 to start my own group!
We will explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology.
PhD positions available!
12.10.2025 20:19 — 👍 74 🔁 30 💬 6 📌 5

Join us for the Austrian Cryo-EM Symposium from Nov 11–12 at ISTA! Top speakers, cutting-edge cryo-EM, and a chance to explore Vienna & the ISTA campus. Register now 👉 cryoem-symposium.pages.ist.ac.at/registration/
#cryoEM
10.10.2025 10:04 — 👍 17 🔁 8 💬 1 📌 0
... or a 30S proteasome.
11.10.2025 22:37 — 👍 1 🔁 0 💬 0 📌 0
The last one looks like a 26S proteasome (~350Å long) to me. The one before that also seems to be a proteasomal assembly. Hard to say which one but could be a hybrid proteasome (19S one one side of the 20S and PA200 or PA28 on the other side).
11.10.2025 22:36 — 👍 2 🔁 0 💬 2 📌 0

A good day to remember John Gurdon’s school report from his biology master at Eton
07.10.2025 20:45 — 👍 79 🔁 19 💬 3 📌 5
Laura Lorenzo Orts
IMB Mainz
I am excited to announce that I will be moving to IMB Mainz next year! The Winter call for the IPP PhD program is now open; if you are interested in maternal #mRNA regulation and #translation in early vertebrate development, please apply! Deadline: 16 October.
More info: www.imb.de/students-pos...
15.09.2025 12:13 — 👍 65 🔁 25 💬 3 📌 3
Makes sense, thanks for the explanation 🙂
Looking forward to play around with it once available on EMPIAR. It seems ideal to get a grasp on the processing of (pseudo)symmetric complexes with symmetrie breaking interaction partners.
02.09.2025 09:13 — 👍 1 🔁 0 💬 0 📌 0
I was surprised to see that the classification was that complicated. Did you try refinement and 3D classifications with or without image alignment in C1 before refining in C16?
Either way this is such a cool processing problem resulting in such a cool structure and story.
Much congrats!
01.09.2025 19:11 — 👍 5 🔁 0 💬 1 📌 0
Glad to share the final version of our story about the UBR4 complex, an E4 ligase protein quality control hub @science.org. Now with more cryo-EM structures and a deeper dive into substrate recognition, especially escaped mitochondrial proteins @clausenlab.bsky.social www.science.org/doi/10.1126/...
28.08.2025 18:54 — 👍 80 🔁 30 💬 7 📌 1
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
27.08.2025 16:14 — 👍 304 🔁 109 💬 14 📌 11

If you are thinking about the future of structural biology, we are too! Our two cents (Jürgen Plitzko and I) just got published online here: authors.elsevier.com/sd/article/S...
23.08.2025 23:21 — 👍 89 🔁 27 💬 2 📌 0
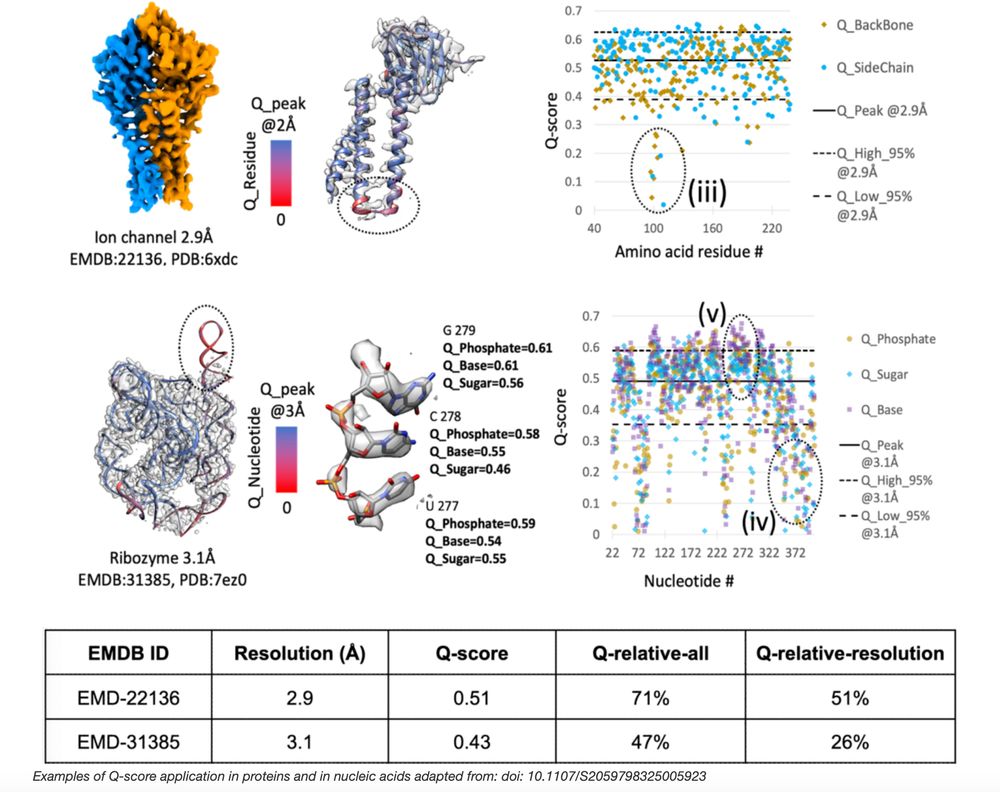
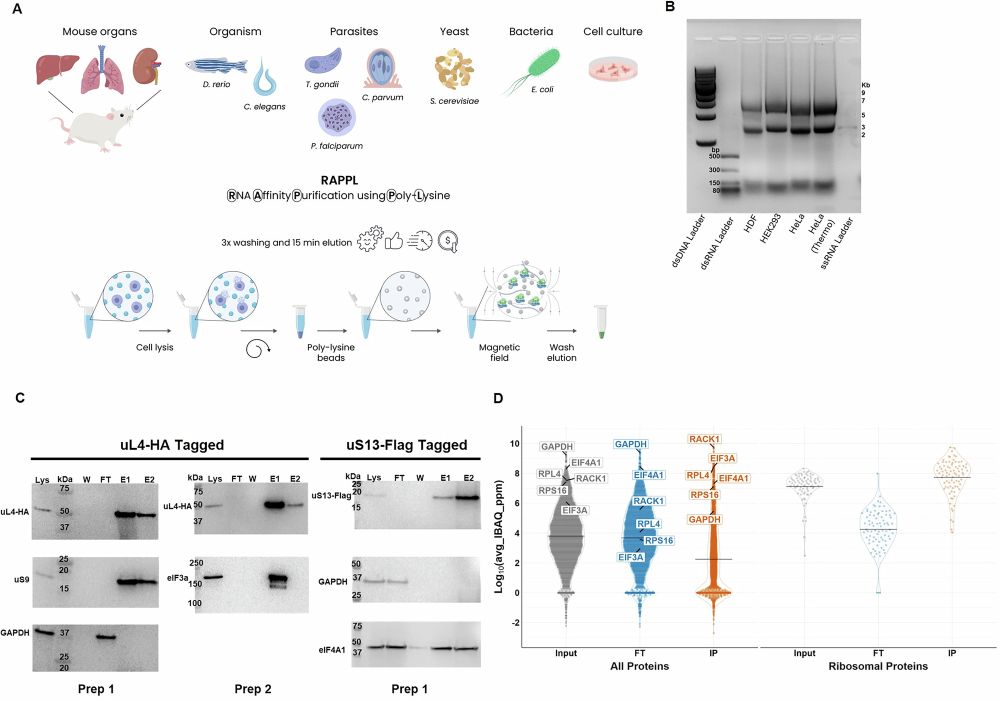
📄 New paper: 3DEM Structure Map Validation Recommendations
Two new metrics assess model-to-map fitness in cryo-EM:
🔹 Q_relative_all → compares avg Q-score across PDB/EMDB
🔹 Q_relative_resolution → compares within similar resolution
#3DEM #CryoEM #wwPDB
20.08.2025 12:53 — 👍 21 🔁 12 💬 1 📌 0

We’re hosting the @embo.org Practical Course on In-situ structural biology by CryoFIB and CryoET in April 2026! 🧬❄️ You can expect hands-on training, cutting-edge methods, and a chance to work alongside leading experts in structural biology. 🔗 Apply by Dec 8, 2025: meetings.embo.org/event/26-in-...
18.08.2025 11:29 — 👍 31 🔁 18 💬 0 📌 2

Integrated structural biology of the native malarial translation machinery and its inhibition by an antimalarial drug
Nature Structural & Molecular Biology - Integrated structural biology approach leveraging in situ cryo-electron tomography reveals molecular details of the native malarial translation...
Our #CryoET story on how a top antimalarial drug candidate perturbs the native malarial translation machinery is out @natsmb.nature.com🥳 rdcu.be/eBbrH
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
18.08.2025 12:13 — 👍 87 🔁 38 💬 4 📌 2
Very nice, congratulations "Danger" Daniel 😆
07.08.2025 06:41 — 👍 1 🔁 0 💬 1 📌 0
Come join us for the Austrian Cryo-EM Symposium!
27.07.2025 08:11 — 👍 1 🔁 0 💬 0 📌 0
That's a wrap! The results of the first #cryoEM heterogeneity challenge are up on biorxiv!
biorxiv.org/content/10.110
23.07.2025 21:43 — 👍 45 🔁 22 💬 3 📌 4
Very cool, congratulations! I was waiting for this 😄
Seems like I will have to compare my favorite methods 🎉
24.07.2025 16:39 — 👍 1 🔁 0 💬 0 📌 0
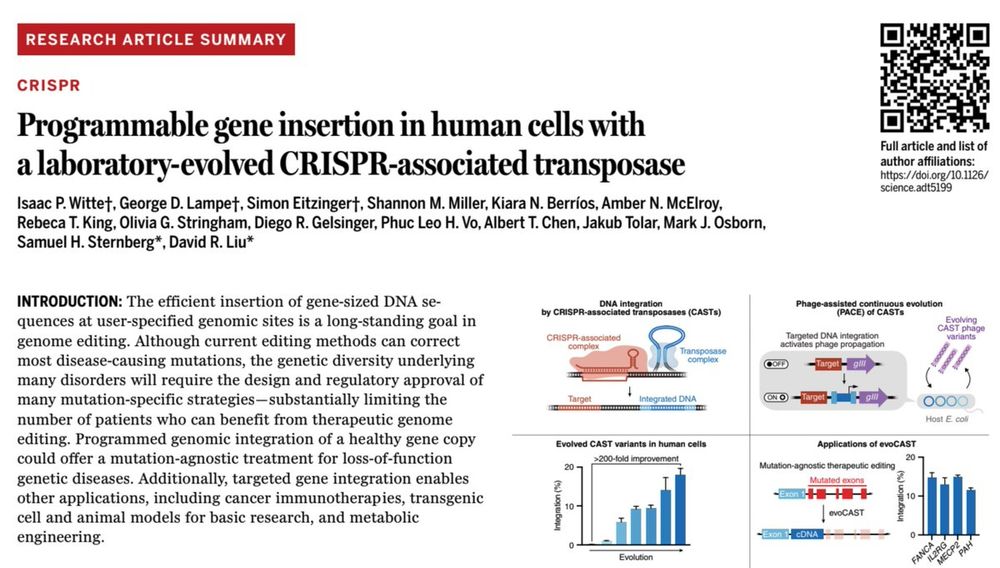
1/10 Today in @science.org in collaboration with
the Liu group we report the development of a laboratory-evolved CRISPR-associated transposase (evoCAST) that supports therapeutically relevant levels of RNA-programmable gene insertion in human cells. drive.google.com/file/d/1I-Ub...
15.05.2025 22:49 — 👍 132 🔁 59 💬 2 📌 5

Congrats to @lgrundmann.bsky.social for winning a poster award at #EMBORNAmeetsproteindecay
15.05.2025 05:51 — 👍 18 🔁 2 💬 1 📌 0
The 'RNA meets Protein Decay' @embo.org workshop was really a blast. Thanks to everyone for the lively discussions!
I am especially honoured that my poster was awarded with one of the two poster prizes 😇
15.05.2025 19:25 — 👍 7 🔁 1 💬 0 📌 0


The 2nd edition of the @embo.org workshop 'RNA meets Protein Decay', this time here in Vienna @impvienna.bsky.social, came to an end. Thanks to everyone who came and contributed to the great vibe of sharing science, discussing ideas and connecting with each other!
15.05.2025 10:46 — 👍 26 🔁 3 💬 2 📌 4
Prof for chemical biology at ETH Zurich
Interested in: genetic code expansion - chembio tools - bioorthogonal chemistries - PTMs - Ub in all its shades - protein engineering
Senior Scientist at ETH Zurich. Developing GCE tools to dissect complex biological Systems.
Revealing the hidden world of shape shifting enzymes using Cryo-EM. Senior research associate at Newcastle University, UK. Currently working as part of Recon4IMD https://www.recon4imd.org Views are my own.
The Institute of Science and Technology Austria (ISTA) is dedicated to cutting-edge basic research and graduate education.
www.ista.ac.at
We study how organisms grow, age & develop disease through basic research in epigenetics, genome stability & related fields 🧬
www.imb.de / Imprint: https://www.imb.de/impressum-imprint
📍 #Mainz, Germany
Senior Group Leader IMP / Adjunct Professor Medical University of Vienna. Fascinated by functional genetics (CRISPR, RNAi, degrons) and time-resolved omics towards understanding cancer biology and probing new therapeutic concepts.
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan, Board of Directors NumFOCUS
Previously IMP Vienna, Stanford Genetics, UW CSE.
Briggs group at MPI Biochemistry
Department of Cell and Virus Structure
Cryo-EM, tomography, CLEM, coated vesicles, enveloped viruses.
Operated by @shervinnia.bsky.social and inspired by the now-defunct bot by Frédéric Poitevin and @gati.bsky.social. Suggestions welcome.
Asst Prof at ISTAustria. Lover of tiny things including proteins, tardigrades, seashells & round furry animals.
mRNA & cryo-EM enthusiasts at IMP Vienna. Posts are by lab members.
Scientist @arcinstitute
Ex–@stark_lab, @IMPvienna, @viennabiocenter
I like molecular mechanisms, music, and evolutionary biology | PhD student in the Burga lab at IMBA, Vienna
PhD student at the Brennecke and Plaschka labs, Vienna.
Interested in RNA silencing
🇨🇦Postdoc in the Burga lab at the Institute of Molecular Biotechnology (IMBA)🇦🇹. Developmental biologist turned selfish gene/mobile genetic element/virus enthusiast, working in nematodes at the interface of them all!
Assistant Prof and Group Leader at Max Perutz Labs, University of Vienna
Computational Cryo-EM/ET at Genentech
Love people, the outdoors and building stuff
Complexity is the enemy
All opinions my own
The national center for inter-university research facilities at Seoul National University.
Cryo-EM / Structural Biology
CEO Puxano/CryoEM&ProteinDesign method development at the exp. and comp. level/Open Science initiative supporter/impact entrepreneurship/ art (philosophy)
Views expressed are my own.
Marie Curie Postdoc Fellow at Utrecht University 🇳🇱 Protein-Biochemist diving into cryo-ET ❄️