🔥my latest paper from @odedrechavi.bsky.social lab🔥
we found small RNAs act across tissues to regulate fertlity in C. elegans 🪱. Surprisingly, we also found that O2-sensing neurons inhibit germline maintenance. Follow along our journey👇https://www.biorxiv.org/content/10.1101/2025.08.07.669182v1
11.08.2025 08:26 — 👍 56 🔁 27 💬 8 📌 1
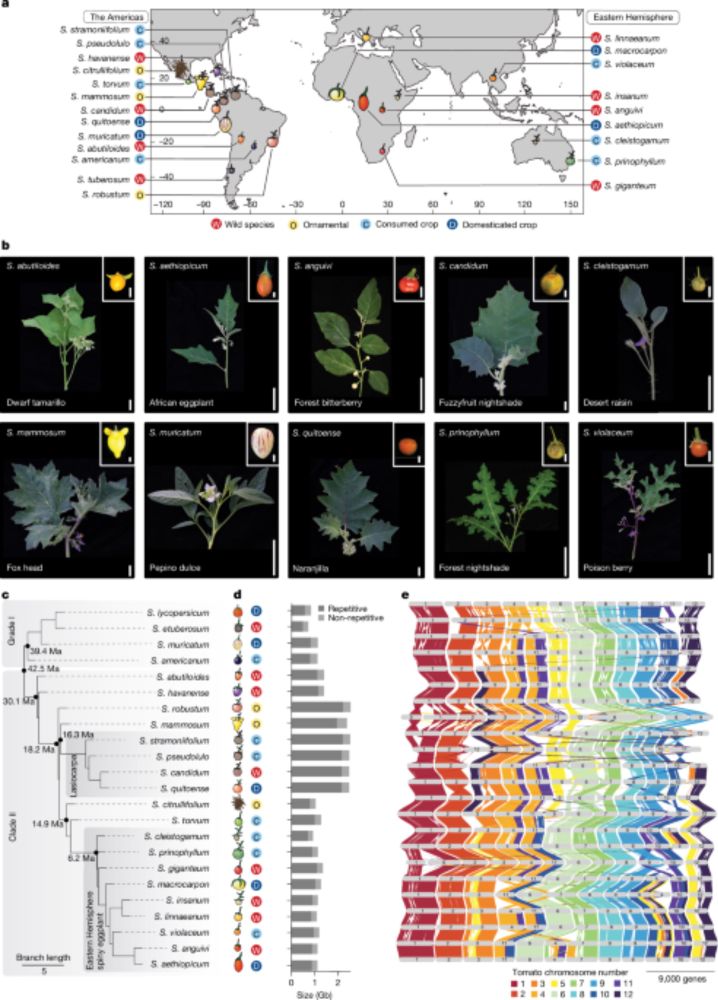
Solanum pan-genetics reveals paralogues as contingencies in crop engineering - Nature
Gene duplication and subsequent paralogue diversification are major obstacles to genotype-to-phenotype predictability.
🚨New paper published in @nature.com! Using pan-genetics across the Solanum genus🍅🥔🍆we reveal why gene duplications🧬are major contingencies in crop engineering. My postdoc work in the Lippman lab @CSHL, collab. with @katiejenike.bsky.social @mikeschatz.bsky.social chatz.bsky.social and many others!
05.03.2025 16:43 — 👍 194 🔁 104 💬 18 📌 7
we believe our findings are largely consistent with macroevolutionary theories of transcriptome evolution - this suggests that gene regulatory networks might evolve in similar ways across time scales
26.02.2025 01:17 — 👍 0 🔁 0 💬 1 📌 0
key takeaways
⚖️ selection is mostly weak - drift dominates!
🕸️ core genes face stronger stabilizing selection (but trans-regulatory variation can relax this)
🧠 tissue-specific genes are under stronger selection - with neuronal and reproductive tissues evolving the fastest
26.02.2025 01:17 — 👍 0 🔁 0 💬 1 📌 0
what we did:
🧬 we used genotypic selection analyses to link variation in transcript levels to fecundity to test how natural selection shapes transcriptomes within a single generation
🕸️ we then integrated our models with network models to explore what constrains/drives transcriptome evolution
26.02.2025 01:17 — 👍 0 🔁 0 💬 1 📌 0
Pseudo-Physicist | PhD student | Playing with Gravitational-Wave detectors @ UC Riverside | Linux Enthusiast | Mustache-bearer | Runner | Feminist | 🇪🇺🏳️🌈
Global change ecology | remote sensing | ecological modeling
Research scientist/adjunct professor @ SDSU Geography
Professor passionate about student success & team science #firstgen #Brassica enthusiast walking Dogs of the Plant World
#Agriculture #Food #HigherEd #EduSky #Agsky #SciComm & more
#Soil #Crop #Science ; opinions mine
Plant Biologist @LIPME Toulouse & Chargé de
Recherche @INRAE | Genome plasticity, chromatin biology & epigenetics in plant-microbe symbioses | He/Him
Professor of Phytopathology | Kiel University
| Crop wild relatives and their pathogens | Likes quantitative and qualitative resistance mechanisms & popgen
Postdoc, firstgen, scientist, worm breeder, cats dad 🏳️🌈
WALII is an NSF-funded Biology Integration Institute focused on understanding anhydrobiosis - how life continues without water.
Register for our first inaugural symposium: https://www.walii.science/virtual-symposium.html
organismal biologist & geneticist | PI at med school in Houston | amazed by microbial superpowers | he/him
#microbiome #Celegans #spacebiology #firstgen 🧫 🦠 🧪 🧬
GS: https://bit.ly/goog-schol-buck-sam
ORCID: 0000-0002-4347-3997
Cutting-edge research, news, commentary, and visuals from the Science family of journals. https://www.science.org
Or just “Li” |
Assist. Prof. @ Plant Bio Michigan State U. |
Also post data visualization |
Lab: https://cxli233.github.io/cxLi_lab/ |
GitHub: https://github.com/cxli233
SMBE fosters communication among molecular evolutionists and advances the field.
🔗 smbe.org
💬 smbe2026.org • biosig.lab.uq.edu.au/strphy26
👥 @smbe-idea.bsky.social
📙 @molbioevol.bsky.social • @genomebiolevol.bsky.social
Postdoc @UCRiverside. Interested in post-transcriptional/translational regulation, and plant adaptation.
Scientific Director of the VIB-UGent Centre for Plant Systems Biology, Professor at Ghent University, the University of Pretoria (SA), and Nanjing Agricultural University (China). Hate migraines and loves Alfie.
Our research group is interested in Chemical Ecology, Natural Product Chemistry, Organic Chemistry, arthropods, e. g. spiders, butterflies, or Collembola, frogs, bacterial volatiles and communication, as well as plants. We are located in Braunschweig, D.
Biologist
A little obsessed by aphid galls
Run a little lab @hhmijanelia.bsky.social
Moving to @stowersinstitute.bsky.social Feb 1, 2026
Provide unsolicited advice to improve your science (or any) talk at howtogiveatalk.com
Scientist@RIKEN, Univ of Tokyo,
Studying plant-microbe interactions, parasitic plants, plant immunity
http://plantimmunity.riken.jp/index.html
Biologist, systems biology, C. elegans, metabolism, gene regulation, networks, horses, cats, (still) happy in academia, UMass Chan Medical School
https://www.umassmed.edu/walhoutlab/
Opinions my own
Assistant Professor of Evolutionary Systems Biology
@UCRiverside, #FirstGen Academic, evolution of interactions between parasitic nematodes and their hosts, including plants and insects
PhD candidate @ UC Riverside in Botany and Plant Sciences 🌿
Cellular and Molecular biologist 🧫
Scholar activist📚
PA✈️CA


