thanks Sarah!! We are excited to try it out with an expert experimental biophysicist if someone is interested 😀
18.11.2025 15:15 — 👍 0 🔁 0 💬 0 📌 0thanks Sarah!! We are excited to try it out with an expert experimental biophysicist if someone is interested 😀
18.11.2025 15:15 — 👍 0 🔁 0 💬 0 📌 0
Open science tools like Simularium help bring science to life.
@mejohnson81.bsky.social shares how it has helped her stoke curiosity in @jhuartssciences.bsky.social students and work with colleagues across timezones.
#OpenScienceWeek
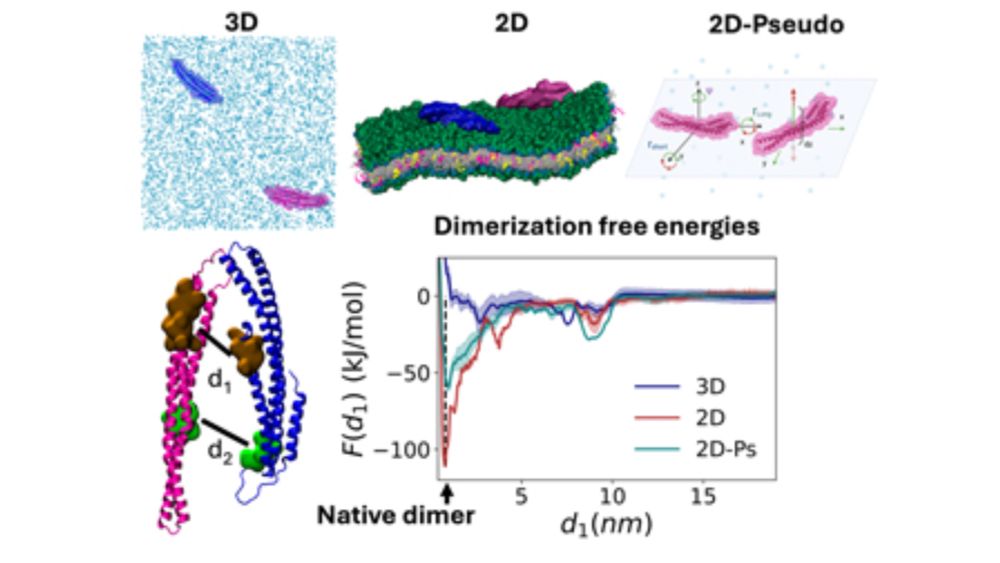
How much do dimer binding affinities change for proteins on the 2D membrane vs in solution? We go beyond rigid-body entropy estimates to show conformational variations can strongly select for 2D association. Very proud of Dr Adip Jhaveri and team's comprehensive work! doi.org/10.1063/5.02...
21.07.2025 21:53 — 👍 6 🔁 0 💬 0 📌 0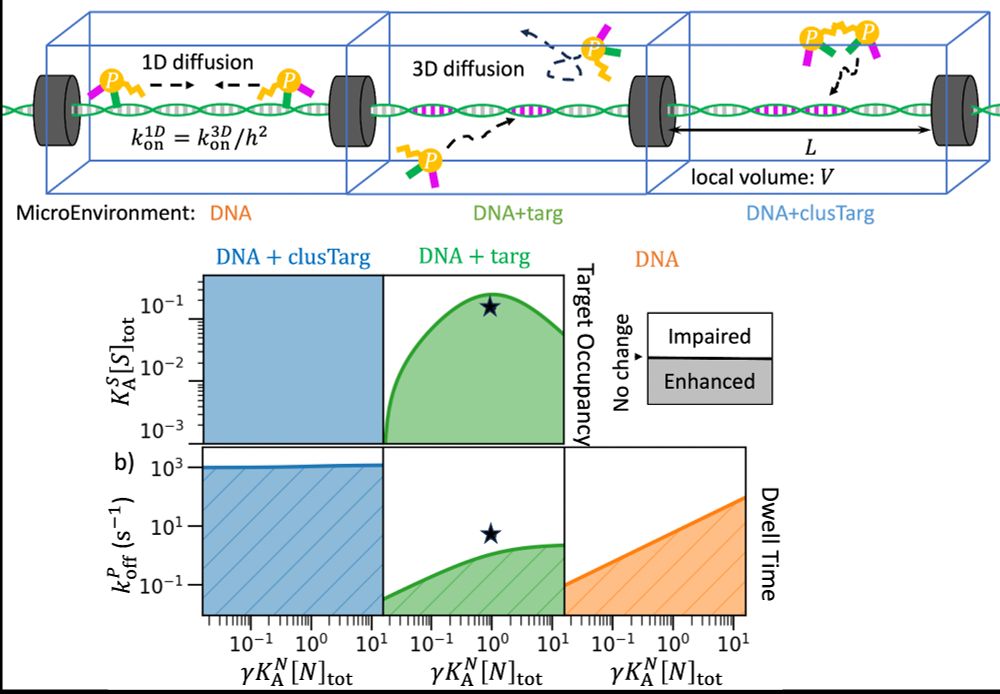
Proteins can form dimers and bind DNA specifically and nonspecifically, exploiting 3D and 1D diffusion. This can drives both enhanced and impaired dwell time and target occupancy depending on spacing between specific targets.
Does protein dimerization always help targeting and dwell time on DNA? Not for a population! Dwell time is especially sensitive to spacing between DNA targets. Nonspecific binding and facilitated (1D) diffusion also play a key role. Super proud of Mankun Sang’s work!
doi.org/10.1101/2025...
The Natural Philosophy Symposium is underway! I had some idea of live-skeeting, but the real-time event is too absorbing.
www.naturalphilosophyhopkins.org/natural-phil...
Anyway, we've had:
(cont)
Can sensing of membrane receptors be switched on at tunable threshold densities, without active processing? Proud to share this preprint led by the awesome Dr Foley, please check it out!
27.05.2025 16:17 — 👍 15 🔁 3 💬 0 📌 0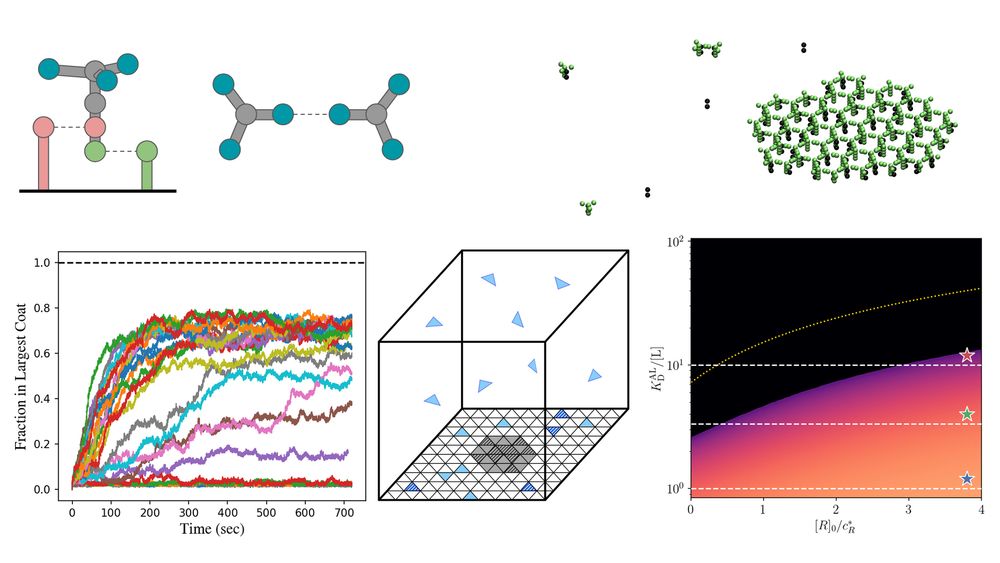
Coarse-grained reaction-diffusion simulations! Idealized lattice models! Coat assembly phase diagrams! Stop on by for a theoretically fun time!
For any of my friends at the #APS Global Physics Summit, come to the Macromolecular Assembly in Cells II session today at 3pm to hear from me about how clathrin self-assembly is tuned to sense cargo receptors!
19.03.2025 15:41 — 👍 4 🔁 2 💬 1 📌 0