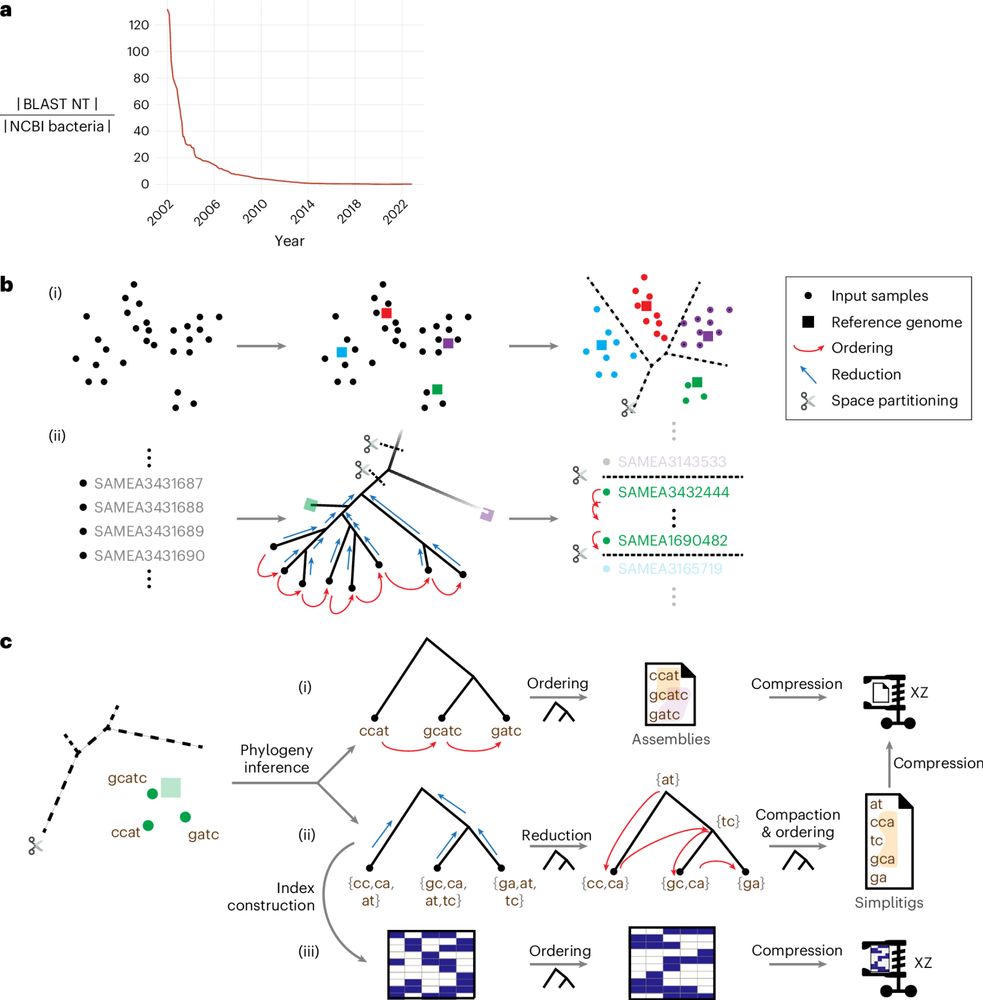
How did @Nature become "prestigious" to scientists?
In our opening article of Issue 09, writer Robert Reason traces the journal's history.
By understanding how Nature’s prestige was constructed, we can also clarify which elements are deserved and which are entrenched.
05.01.2026 16:14 —
👍 35
🔁 16
💬 1
📌 2
Our new paper on high-throughput isolation of Bifidobacterium just got published! A very nice adventure starting with the supervision of @lamhaiha.bsky.social, a talented intern, and a nice collab with our BSight expert @isaacyueyuan.bsky.social! Stay tuned for more isolation and Bifido work! :)
01.01.2026 09:20 —
👍 7
🔁 2
💬 0
📌 0
Hi Sean! Looks very interesting! Will the talks be recorded and shared in some ways? Sadly it will happen during night time for some of us. Should we still register to get access to the records? Thanks!
28.11.2025 09:23 —
👍 1
🔁 0
💬 1
📌 0

New blog post – A quick look at Roche's SBX
lh3.github.io/2025/09/11/a...
12.09.2025 03:26 —
👍 57
🔁 30
💬 2
📌 3
Preprint out for myloasm, our new nanopore / HiFi metagenome assembler!
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
07.09.2025 23:34 —
👍 114
🔁 80
💬 5
📌 5
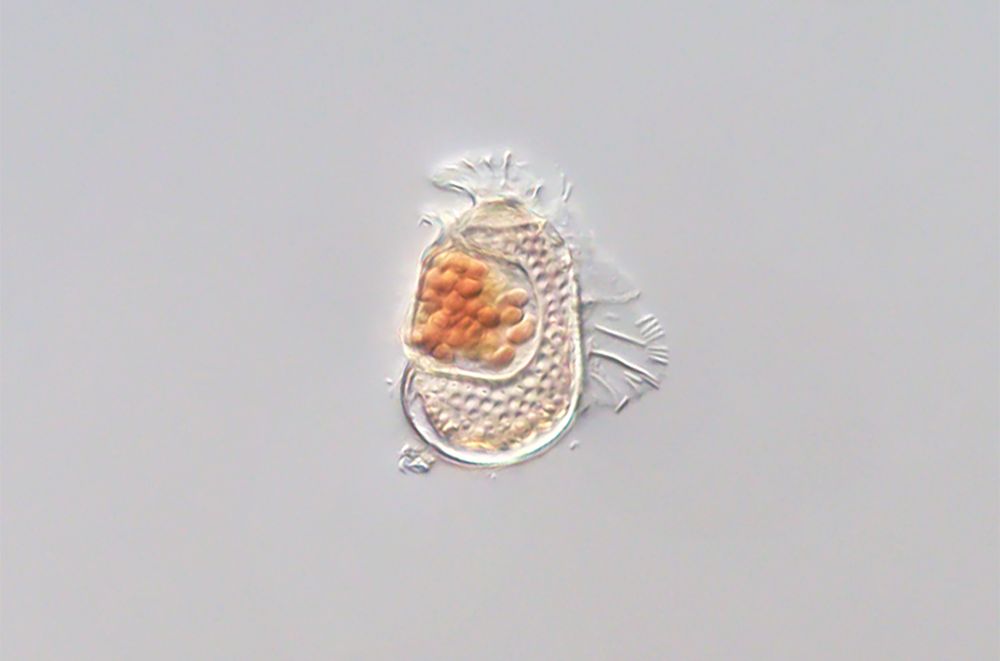
Hidden inside this dinoflagellate is a tiny microbe so reliant on its host that scientists say it’s the most viruslike cellular organism known. Takuro Nakayama/University of Tsukuba
The most viruslike microbe to date was discovered by accident when researchers sequenced the DNA inside this tiny ocean critter. That story and more of the best from @science.org and science in this edition of #ScienceAdviser: www.science.org/content/arti... 🧪
16.06.2025 20:08 —
👍 100
🔁 24
💬 4
📌 1
Short-read metagenomic sequencing cannot recover genomes from many abundant marine prokaryotes due to high strain heterogeneity and platform-inherent GC bias (likely viruses, too), but Nanopore long reads can address this. A results thread on our recent preprint 🧵.
25.05.2025 10:27 —
👍 44
🔁 23
💬 1
📌 4

LLMs can't stop making up software dependencies and sabotaging everything
: Hallucinated package names fuel 'slopsquatting'
LLMs hallucinating nonexistent software packages with plausible names leads to a new malware vulnerability: "slopsquatting."
12.04.2025 22:31 —
👍 1340
🔁 387
💬 33
📌 143

In 1904, a physician took soil he had “impregnated with a living emulsion” of a virulent bacterium, Serratia marcescens, and sprinkled it in front of the UK House of Commons.
This was an early experiment to study how microbes spread through buildings and cause infections...
23.03.2025 15:52 —
👍 8
🔁 2
💬 2
📌 1

Home
A tool for generating consensus long-read assemblies for bacterial genomes - rrwick/Autocycler
New year, new assemblies!
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
31.12.2024 23:43 —
👍 156
🔁 97
💬 2
📌 3
Interactive online show about the microbial world from the Museum of Natural history at Oxford Uni. Cool ideas. www.oum.ox.ac.uk/bacterialwor...
13.12.2024 08:33 —
👍 0
🔁 0
💬 0
📌 0

Image of first page and an illustration from our paper titled "The species as a reproductive community emerging from the past".
A new view of #SpeciesConcepts by @jeannettewhitton.bsky.social and me, clarifying time frames while unifying interbreeding, genealogy, and traits. A general-purpose species should be a reproductive community (bound by cohesive processes) viewed retrospectively (RRCC). doi.org/10.18061/bss...
09.12.2023 19:49 —
👍 77
🔁 28
💬 6
📌 7

Francis Crick Was Misunderstood
The Central Dogma is not a
Another small and interesting historical story from Asimov press. This time about the Watson's central dogma.
06.12.2024 02:44 —
👍 0
🔁 0
💬 0
📌 0
Yes this is the main issue for most of us I guess, but I'll let you know if I think of someone.
20.11.2024 05:58 —
👍 0
🔁 0
💬 0
📌 0
The methodology used in previous paper is relatively easy and would probably only take few weeks or a couple of months top to reproduce. The problem is that previous methods were good enough to observe global patterns, but are subject to context-specific issues. So it's a matter of luck I think.
19.11.2024 01:40 —
👍 0
🔁 0
💬 1
📌 0