
🔄 Inversions predispose to recurrent deletions and duplications in chromsome 15q13.3. 🔁
Using de-novo assemblies of 10 patient-parent trios, we investigated how recurrent copy-number variants (CNVs) in the 15q13.3 locus arise.
www.biorxiv.org/content/10.6...
A brief tour (1/17)
06.03.2026 15:27 —
👍 9
🔁 4
💬 1
📌 0

Time for a thread on our Christmas preprint “Origin and evolution of acrocentric chromosomes in human and great apes”. I had so much fun with this project and paper. It will be hard to summarize in a thread, but I’ll try www.biorxiv.org/content/10.6... [1/21]
02.02.2026 14:58 —
👍 41
🔁 29
💬 1
📌 1

Absolutely thrilled to share the latest work from my lab focused on the variation and evolution of human centromeres among global populations! We assembled 2,110 human centromeres, identifying 226 new major haplotypes and 1,870 α-satellite HOR variants. www.biorxiv.org/content/10.6...
16.12.2025 16:05 —
👍 108
🔁 46
💬 4
📌 2
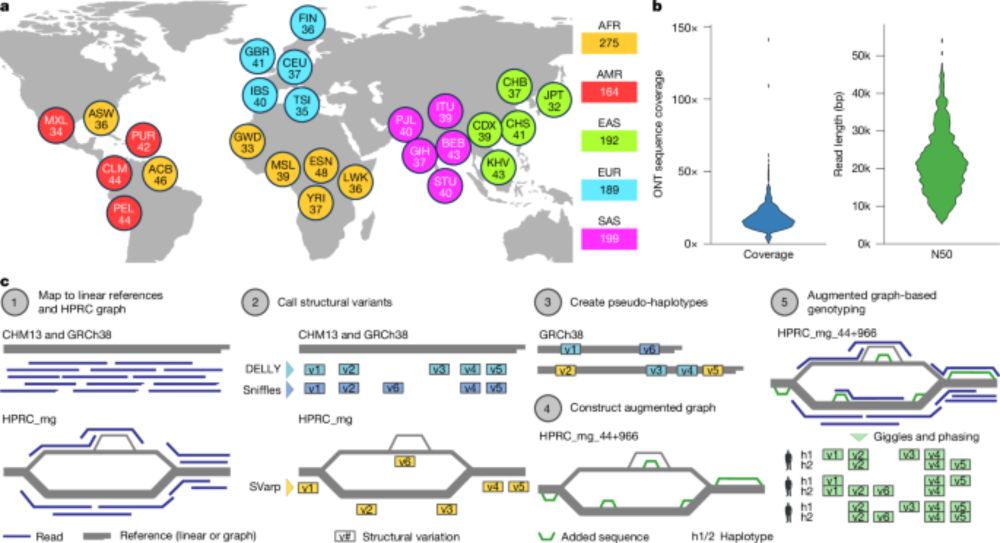
Structural variation in 1,019 diverse humans based on long-read sequencing - Nature
Intermediate-coverage long-read sequencing in 1,019 diverse humans from the 1000 Genomes Project, representing 26 populations, enables the generation of comprehensive population-scale structural varia...
Another taste of the future of human genetics, also in @nature.com. Long-read sequencing in >1,000 humans across 26 populations shows how the approach can accelerate biological & medical research. Work led by @bernardo-rodriguez.bsky.social @trausch.bsky.social @tobiasmar.bsky.social & Jan Korbel.🧬🧪
24.07.2025 10:26 —
👍 14
🔁 6
💬 0
📌 0
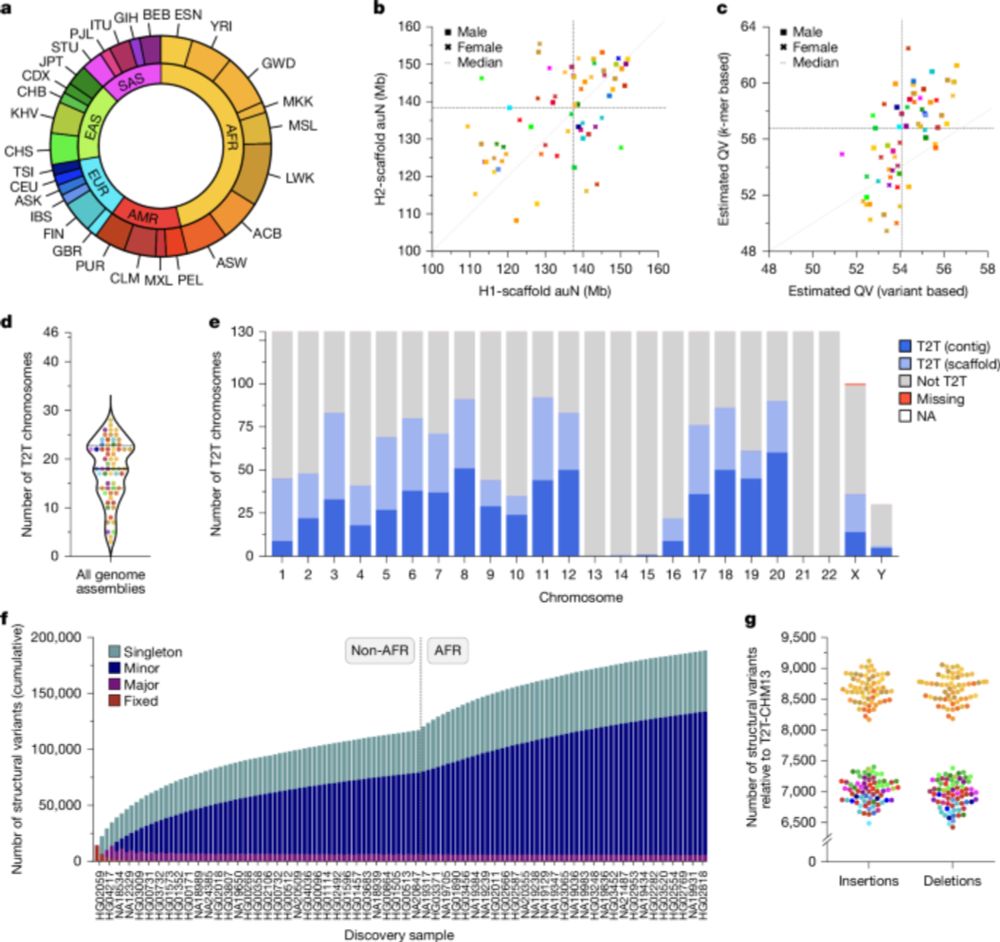
Complex genetic variation in nearly complete human genomes - Nature
Using sequencing and haplotype-resolved assembly of 65 diverse human genomes, complex regions including the major histocompatibility complex and centromeres are analysed.
Telomere-to-telomere DNA sequencing is set to transform the field of human genetics in coming years. For a flavour of what's coming, see this exciting work on nearly complete genomes of 65 individuals from diverse populations, out today in @nature.com by @glennislogsdon.bsky.social & colleagues. 👇🧬🧪
23.07.2025 16:19 —
👍 86
🔁 39
💬 1
📌 1
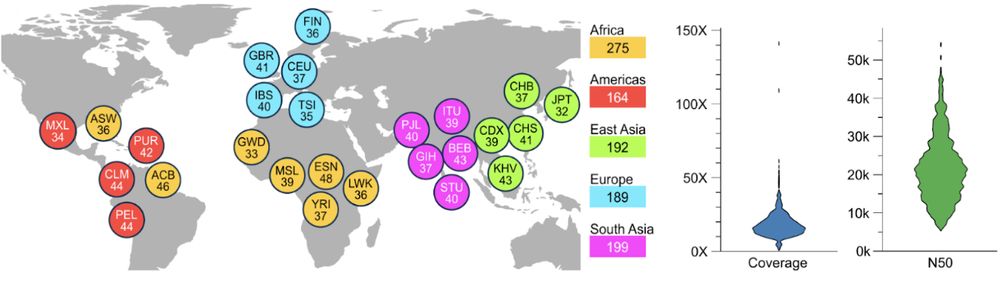
[1/8] *New Open-Access Long Read Resource*. We sequenced 1,019 genomes from the 1000 Genomes Project sample cohort using @nanoporetech.com long-read sequencing (LRS) to median 17x coverage. Publication at go.nature.com/4ffPb8f.
@hhu.de @crg.eu @embl.org @impvienna.bsky.social
24.07.2025 09:26 —
👍 42
🔁 21
💬 1
📌 2
Lovely write up on our paper from @jacksonlab.bsky.social. It's been <4 years since I joined the HGSVC, and together with Peter Audano and Parithi Balachandran in my lab, @tobiasmar.bsky.social, @glennislogsdon.bsky.social, and many others, we've done a lot. Here's to great collaborators!
24.07.2025 01:41 —
👍 9
🔁 1
💬 0
📌 0
I will talk about those papers in my Hitseq keynote tomorrow at #ISMBECCB2025, so if you are in Liverpool don't miss it.
23.07.2025 15:17 —
👍 4
🔁 1
💬 0
📌 0

🖥️🧬 Now, Zahra is talking about oarfish, for your long read RNA-seq quantification needs!
github.com/COMBINE-lab/...
23.07.2025 13:24 —
👍 13
🔁 1
💬 0
📌 0

🧬 & 🖥️
📢Call for Posters #GCB2025 | Join the #bioinformatics and #computationalbiology community at the German Conference on Bioinformatics! Submit your poster now!
Important Deadlines:
📌 Early Bird Registration: 5 July 25
📌 Poster Submission: 7 August 25
🔗 gcb2025.de/GCB2025_call...
02.06.2025 08:43 —
👍 5
🔁 4
💬 0
📌 0

🚨 Registration is OPEN for the 18th Berlin Summer Meeting – Sequencing Planet Earth!
📍 MDC-BIMSB @mdc-berlin.bsky.social
🗓️ June 19–20, 2025
🔗 mdc-berlin.de/BSM2025
#BSM2025 #mdcBerlin #SystemsBiology #Genomics #DataScience
(1/5)
27.03.2025 14:30 —
👍 4
🔁 5
💬 1
📌 0

Submit your abstracts from all areas of #bioinformatics and #computationalbiology for the German Conference on Bioinformatics - #GCB2025 at University of Dusseldorf from 22 - 24 September 2025!
abstract submission: gcb2025.de
@denbi.bsky.social @openbio.bsky.social @SIB.mstdn.science.ap.brid.gy
03.02.2025 13:41 —
👍 12
🔁 9
💬 0
📌 0












