C'est le problème des pauvres, ils sont pas suffisamment disrupteur pour trouver des solutions contre le changement climatique.
02.07.2025 07:30 — 👍 0 🔁 0 💬 0 📌 0
You can now save Ultra HD (8k) resolution screenshot in VIAMD, with or without the GUI. Here illustrated on a visualization of a Moebius ring calculated with @veloxchem.bsky.social. #compchem
01.07.2025 09:44 — 👍 6 🔁 2 💬 0 📌 0

Come and visit us at the @veloxchem.bsky.social booth at #watoc #compchem.
22.06.2025 14:19 — 👍 4 🔁 2 💬 0 📌 0

Road trip to #watoc! See you there. #compchem
21.06.2025 13:15 — 👍 3 🔁 3 💬 0 📌 0
Hi Jean-Philip. Long time no see. Pass by the @veloxchem.bsky.social booth if you have time.
21.06.2025 21:06 — 👍 1 🔁 0 💬 0 📌 0
On my way to the World Association of Theoretical and Computational Chemists (WATOC) meeting in Olso.
#WATOC #CompChemSky #ChemSky
www.watoc2025.no
21.06.2025 09:04 — 👍 19 🔁 2 💬 1 📌 0
I am sorry but the otter is the goat skogsduva.
18.06.2025 14:32 — 👍 0 🔁 0 💬 1 📌 0
See you there. I'll be at the @veloxchem.bsky.social booth. Presenting our QM software as well as @viamd.bsky.social. #WATOC #compchem
18.06.2025 10:11 — 👍 3 🔁 3 💬 0 📌 0

Signez la pétition
Sauvons le Palais de la découverte
Pour la réouverture du palais de la découverte ! chng.it/gncnBRZnTc
13.06.2025 20:48 — 👍 5 🔁 2 💬 0 📌 0
Happy #pride month! We believe that chemistry is for everyone. In the face of opposition that threatens the progress we've made, it's never been more important for us to promote equality of opportunities and make chemistry as open, inclusive and diverse as it should be: buff.ly/1dShs0l #ChemSky
04.06.2025 12:01 — 👍 58 🔁 16 💬 0 📌 2
See you there. I will be at the @veloxchem.bsky.social booth.
04.06.2025 21:33 — 👍 1 🔁 0 💬 0 📌 0
Surprisingly, the UMA (by meta) model captures the bond-breaking process in the carbon nanotube stretching test, similar to GFN2-xTB. I hadn't expected deep learning models to extrapolate over bond lengths.
#compchemsky #chemsky
04.06.2025 15:52 — 👍 6 🔁 1 💬 2 📌 0
Get inside a bacterial cell with this cool combination of @cg-martini.bsky.social and @viamd.bsky.social
Also check out our recent webinars on each:
🎬 Whole cell simulation with Martini ▶️ youtu.be/fvFaPgSoM90
🎬 Visual analysis of #moleculardynamics with VIAMD ▶️ youtu.be/wVENzcx0XmQ
04.06.2025 11:36 — 👍 8 🔁 3 💬 0 📌 1
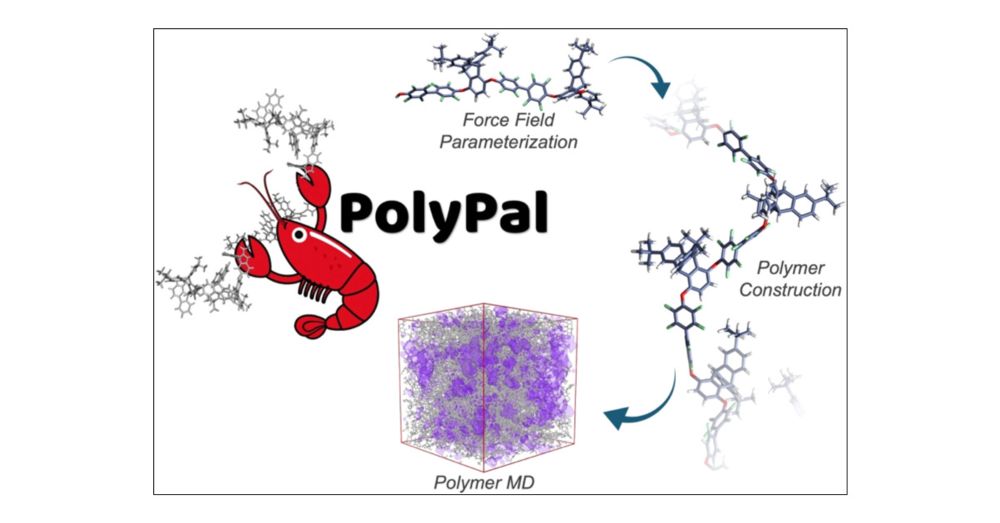
PolyPal: A Python Package for Molecular Dynamics Simulation of Amorphous Polymers
Easily tunable and processable, porous organic polymers (POPs) have found increasing utility in various applications. Molecular modeling and simulations are invaluable tools in polymer science but remain under-reported in the POP literature. Accurate modeling and simulation of these materials could boost the discovery of high-performance POPs and allow for a more thorough contribution to big data. These polymers contain free volume-promoting structural units, such as iptycenes, and exhibit high glass-transition temperatures, excellent thermal stability, and tunable functionality. However, popular transferable force fields utilized in all-atomistic molecular dynamics (MD) simulations are not fully parametrized for intrinsically porous thermoplastic materials. We present a streamlined workflow for all-atomistic MD simulations of nonporous and porous amorphous polymer materials. In conjunction with the programs ORCA, Q-Force, Assemble!, and GROMACS, a highly accessible methodology is established for force field (FF) parametrization, creation of initial configurations, and simulation of various nonporous and porous polymers. This protocol can reproduce experimental bulk densities and fractional free volume values for amorphous polymeric materials with excellent accuracy and has been made available as a Python package, called PolyPal. As an example, we present our results using PolyPal on a series of nonporous and porous polymers that were previously synthesized and experimentally characterized. FF accuracy was also validated through solid-state NMR studies. These simulations will not only open new avenues for the rational design of high-performance POPs through the contribution of improved insight but also provide a streamlined pathway for simulating previously unexplored porous polymeric materials.
Very interesting #compchem article: pubs.acs.org/doi/10.1021/...
04.06.2025 09:17 — 👍 1 🔁 0 💬 0 📌 0
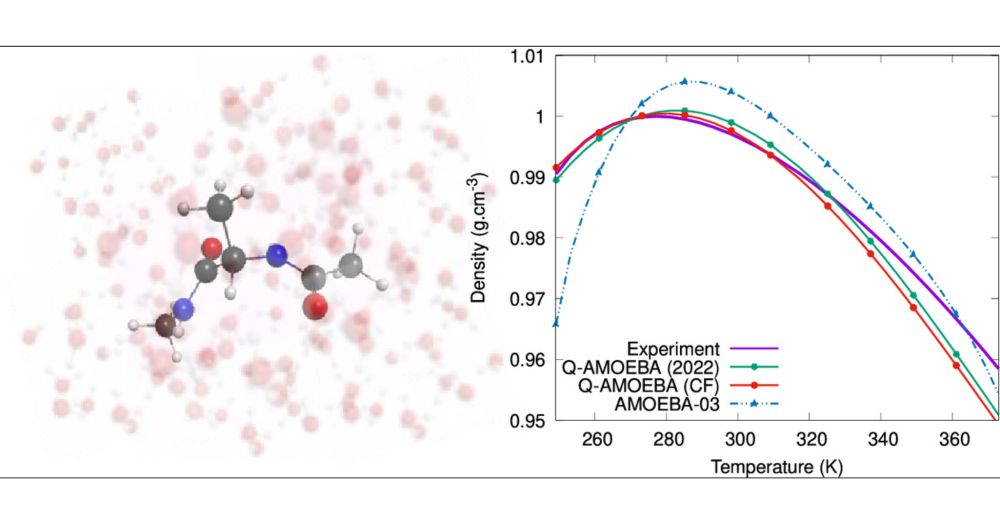
The Q-AMOEBA (CF) Polarizable Potential
We present Q-AMOEBA (CF), an enhanced version of the Q-AMOEBA polarizable model that integrates a geometry-dependent charge flux (CF) term while designed for an explicit treatment of nuclear quantum e...
#compchem New paper in J. Phys. Chem. Lett.
: "The Q-AMOEBA (CF) Polarizable Potential".
Enhanced version of the Q-AMOEBA polarizable model integrating a geometry-dependent charge flux term while designed for an explicit treatment of nuclear quantum effects.
pubs.acs.org/doi/10.1021/...
03.06.2025 20:22 — 👍 8 🔁 4 💬 2 📌 0
MA(R/S)TINI 3: An Enhanced Coarse-Grained Force Field for Accurate Modeling of Cyclic Peptide Self-Assembly and Membrane Interactions | Journal of Chemical Theory and Computation pubs.acs.org/doi/full/10....
03.06.2025 12:04 — 👍 2 🔁 2 💬 0 📌 0
Interactive visualization of @cg-martini.bsky.social bacterial cell with @viamd.bsky.social. #compchem @gromacs.bsky.social
02.06.2025 13:44 — 👍 8 🔁 2 💬 0 📌 1
Thanks. Looking forward to your feedback.
28.05.2025 15:56 — 👍 0 🔁 0 💬 0 📌 0
In addition to our website veloxchem.org, we are now opening the discussion on github, this is the place to exchange and ask questions about veloxchem (github.com/orgs/VeloxCh...). #compchem
27.05.2025 13:04 — 👍 5 🔁 3 💬 1 📌 0
GitHub - VTX-Molecular-Visualization/VTX
Contribute to VTX-Molecular-Visualization/VTX development by creating an account on GitHub.
New publication out!
doi.org/10.1093/bioi...
VTX is open source and freely accessible for non commercial use! github.com/VTX-Molecula...
Builds available at vtx.drugdesign.fr
14.05.2025 15:36 — 👍 14 🔁 2 💬 1 📌 2

Happy to see ChemRxiv now has a Bluesky button! #chemsky
28.04.2025 08:07 — 👍 58 🔁 14 💬 1 📌 0
#compchem, I will be teaching in this workshop. Join if you want to learn about @viamd.bsky.social and @veloxchem.bsky.social
25.04.2025 13:56 — 👍 3 🔁 0 💬 0 📌 0
@viamd.bsky.social @veloxchem.bsky.social
25.04.2025 13:13 — 👍 0 🔁 0 💬 0 📌 0
New feature! You can now visualize trajectories with bond formation and breaking by checking the Recalc Bonds option. #compchem
23.04.2025 09:22 — 👍 1 🔁 1 💬 0 📌 0
The problem with most machine-based random number generators is that they’re not TRULY random, so if you need genuine randomness it is sometimes necessary to link your code to an external random process like a physical noise source or the current rate of US tariffs on a given country.
09.04.2025 19:15 — 👍 19309 🔁 3673 💬 389 📌 249
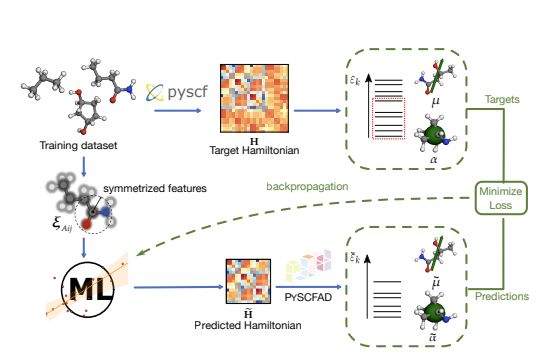
A schematic of the functioning of a ML/QM hybrid framework
When you combine #machinelearning and #compchem, you need to start worrying at the QM details within your ML architecture. We use our indirect Hamiltonian framework and pySCFAD to explore the enormous design space arxiv.org/abs/2504.01187
03.04.2025 21:30 — 👍 14 🔁 4 💬 2 📌 0
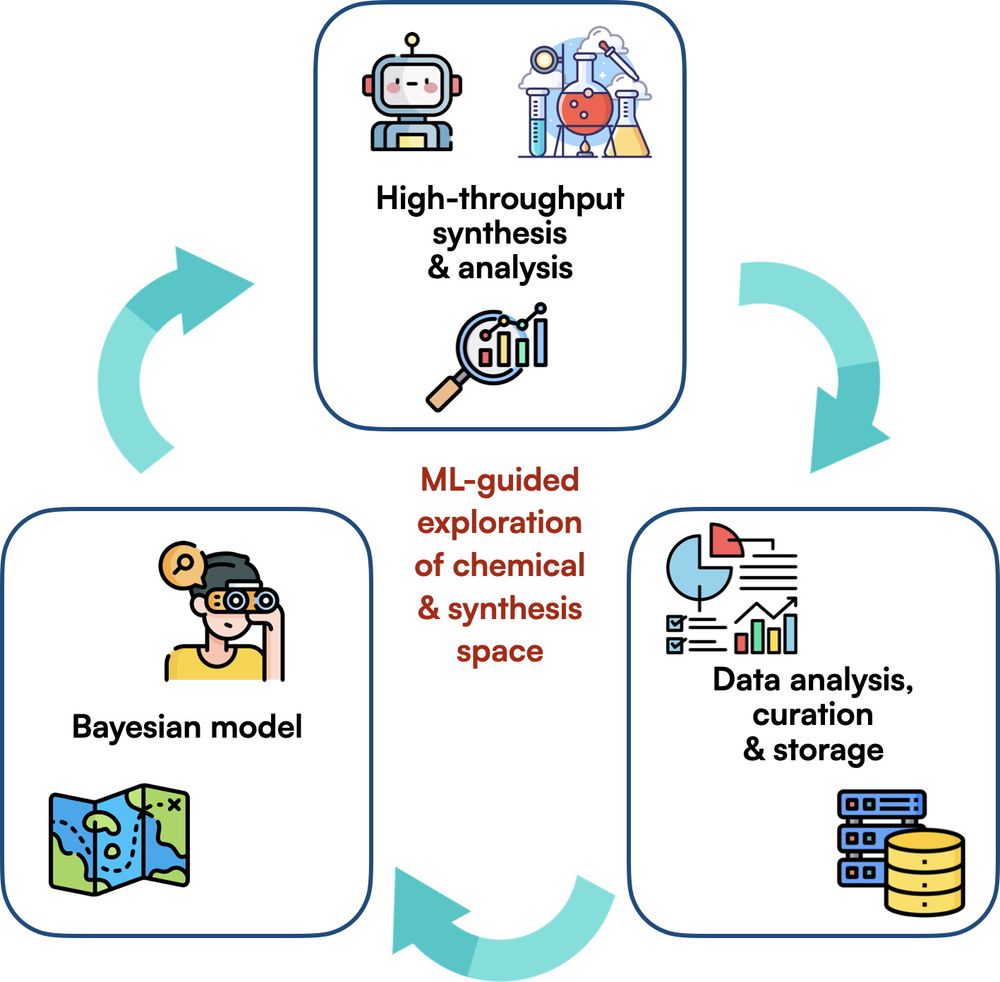
In the center: "ML-guided exploration of chemical & synthesis space".
Around it, three axes of work: "High-throughput synthesis & analysis", "Data analysis, curation & storage", "Bayesian model".
📢 PhD offer in my group: “Active learning to accelerate discovery of multi-variate MOFs” — I'm very excited about this new project, #compchem with a strong collaboration with experimental group of Christian Serre / IMAP #chemsky
🔗 www.coudert.name/tmp/PhD_posi...
07.04.2025 08:48 — 👍 50 🔁 33 💬 1 📌 0
Senior Research Prof. IMDEA Nanoscience Madrid
💻🧪Photophysics of Organic & Hybrid Materials
😐💪 Scientific Integrity Decline
Profile: nanoscience.imdea.org/home-en/people/item/johannes-gierschner
Website: www.uv.es/jogiers
orcid.org/0000-0001-8177-7919
Computational Biochemistry | CovinoLab
PhD in Chemistry @UniBarcelona | Computational Chemistry | Molecular modeling | Enzyme catalysis | Glycobiology |
Comp chemist, experience modelling reactivity and catalyst design. 💻
Figuring out what's next while brewing coffee... ☕
PhD | Uni of St Andrews | Bühl group 🏴
Scientific Editor, RSC. Views my own.
Background in computational chemistry and education.
Sci-fi, blackgaze, queer horror books, MMOs - including an insistent love of Phantasy Star Online 🏳️⚧️
B. Ch. E.
PhD Theoretical Physicalchemistry
From Mexico to the world! 🇲🇽
Expérimentateur sans blouse blanche. Enquêteur sans moustache. Analyste sans PowerPoint. Vidéaste aussi.
As a division of @acs.org, ACS Publications is proud to publish the most trusted, most cited, and most read collection of journals in the chemical and related sciences.
Explore ACS Publications: pubs.acs.org
recovering academic & development editor at the Royal Society of Chemistry for the journals @pccp.rsc.org @greenchemistry.rsc.org & @faradaydiscussions.rsc.org 🧪 Edinburgh 📌 they/he 🏳🌈🏳️⚧️ big fan of n95 masks & clean air 😷 opinions my own
Multiscale molecular dynamics, biological membranes interactions, CNRS researcher, group leader at @cbitoulouse.bsky.social
#CompChem #Radiochem at the University of Iowa. Loves cats and avoiding the news. UMN Alum. Former Air National Guard member.
Scientist working at the University of Bremen (Faculty of Geosciences)
#mineralogy #crystallography
#zeolites #compchem #adsorption
Assistant Prof. @UvA | amyloids, multiscale soft matter, computational peptide design, smart (adaptable) biomaterials, CompChem
The twitter-er formerly known as @physicsteo. Recovered #CompChem-ist. Doing publishing stuffs @ AIPP. Brooklynite from 🇮🇹 via 🇪🇸, 🇸🇪 & 🇩🇪.
http://matteo.cavalleri.eu
Theoretical Chemistry research group @lct-umr7616.bsky.social, Sorbonne Université & CNRS| Led by Prof. Piquemal (@jppiquem.bsky.social)|
#compchem #HPC #MachineLearning #quantumcomputing
Website: https://piquemalresearch.com
Computational Organic Chemist
@University of Manchester #CompChem 💻 ⚛️ #Catalysis
🇪🇸 → 🇬🇧 | Mum of 3 boys 🤩
Cyclist 🚴♀️ | Yogi 🧘♀️ | Amateur cook
Rock music fan 🎸
🔗 https://trujilloresearchgroup.com/
Professor #compbiophys #compchem @UT_Dallas
QM/MM force fields DNA rep./modif., cancer biomarkers, ionic liquids, #SACNAS, views my own, he/él 🇲🇽🇺🇸
#compchem | organic semiconductors | functional biomaterials | polymers | @mdanalysis core dev | made in Sardinia 🏳️🌈 she/her
Computational chemistry and AI for drug design in biotech #compchem | CSIRO, Melbourne AU