ops 100-300 kya* 😅
01.04.2025 01:16 — 👍 0 🔁 0 💬 1 📌 0ops 100-300 kya* 😅
01.04.2025 01:16 — 👍 0 🔁 0 💬 1 📌 0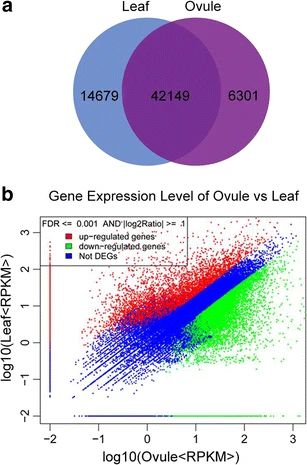
Venn diagram of Gingko genes expressed uniquely in leafs (14679), uniquely in ovules (6310), or expressed in both (42149).
The genomes of Roots and Relicts help understand the evolution of sibling lineages. Wang et al 2016 looked at gene expression for ovules and leaves in Ginkgo and find common mechanisms in genes expressed in the ovule related to flowering genes in angiosperms #RIP #2025MMM doi.org/10.1007/s112...
01.04.2025 01:15 — 👍 17 🔁 3 💬 0 📌 0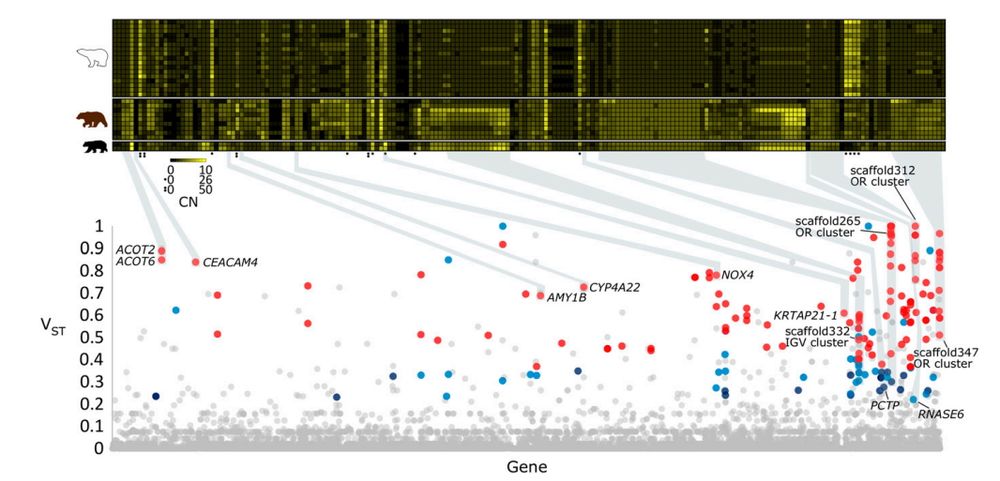
Genes with differences in copy number between polar bears and brown bears. (Lower panel) VST values, which estimate the proportion of population-level copy number variation due to differences between subpopulations in copy number variation, for all 21,142 genes in the polar bear genome. Dots are colored such that red dots correspond to genes with VST > 0.35 in two methods for copy number estimation, purple dots correspond to genes with VST <= 0.35 and VST > 0.22 in two methods for copy number estimation, blue dots correspond to VST > 0.22 in one method for copy number estimation and VST > 0.35 in the second method, and gray dots correspond to VST <= 0.22 in one or both copy number estimation methods. (Upper panel) Heat map of diploid gene copy number for genes (columns) whose copy number is highly differentiated between polar bears and brown bears (n=197 genes). Each row corresponds to individual polar bears (top set of rows), brown bears (middle set of rows), and black bears (bottom set of rows). Yellow corresponds to high copy number and black corresponds to a copy number of 0.
Polar bears and brown bears are close relatives but have very different diets! Polar bears (which eat mostly seals) have fewer copies of the gene AMY1B (important in starch digestion!) relative to brown bears (which consume plants for >70% of their diet). #2025MMM doi.org/10.1073/pnas.1901093116
01.04.2025 00:50 — 👍 25 🔁 4 💬 1 📌 0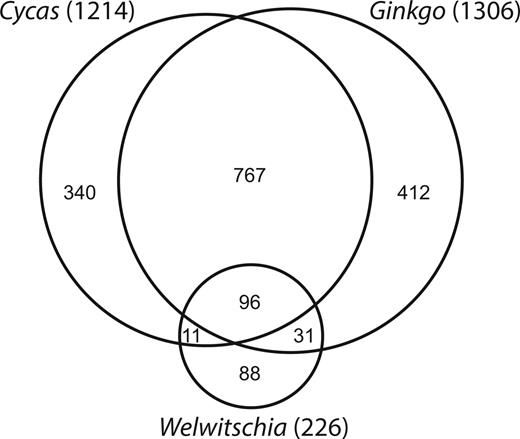
Venn diagram depicting the numbers of shared and unique edit sites in three gymnosperm groups, Cycas, Ginkgo, and Welwitschia
Guo et al 2016 sequenced the ginkgo mitochondrial genomes to look at common features of gymnosperms. The Ginkgo mitogenome is relatively small, have low substitution rates, and possess numerous genes, introns, and edit sites; all present in the ancestral seed plant. doi.org/10.1093/molb... #2025MMM
01.04.2025 00:46 — 👍 20 🔁 7 💬 0 📌 0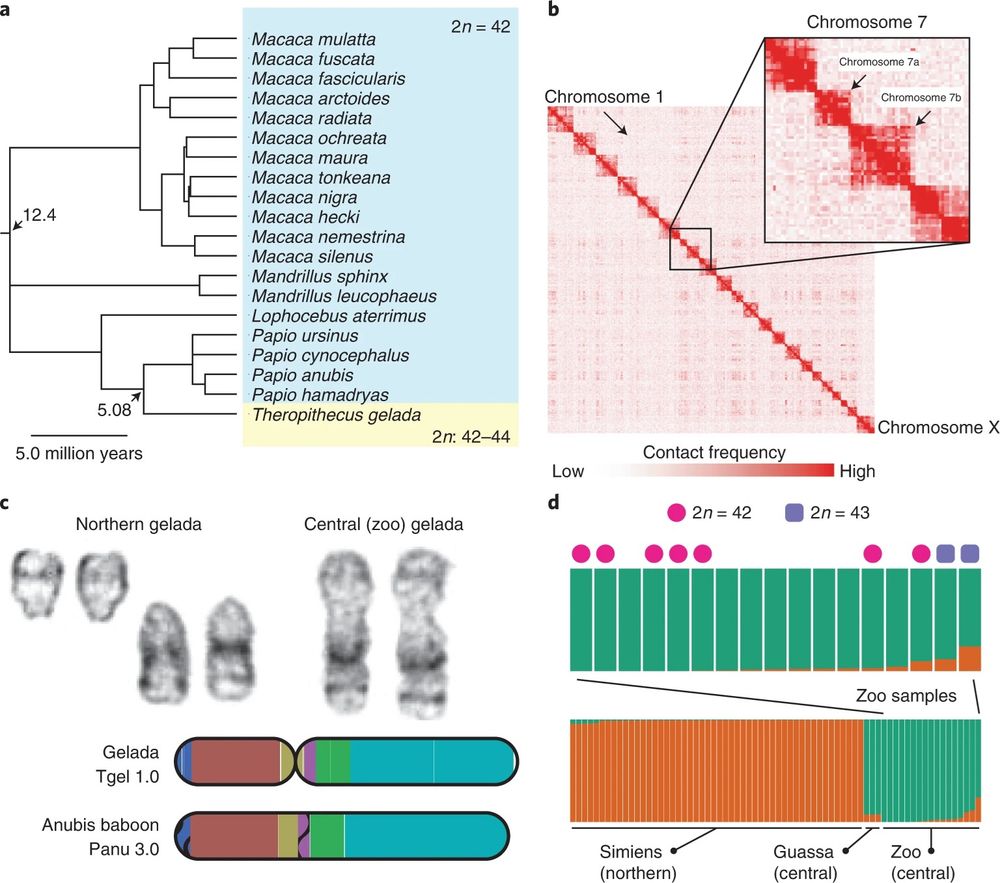
Fig. 2 from Chiou et al. (2022) a, Apart from geladas, the papionin clade exhibits an extremely conserved karyotype of 42 diploid chromosomes. Twenty species with known karyotypes sampled by Stanyon et al.24 are shown with the consensus chronogram from TimeTree53,54. b, Hi-C contact map reveals a distinct lack of contacts between the arms of chromosome 7 in the reference gelada female. c, G-banded karyotyping and analysis of genomic rearrangements reveal strong synteny between fissioned chromosomes and the intact arms of chromosome 7 in central geladas and baboons, respectively. d, STRUCTURE-style bar plot showing ancestry proportions from 70 resequenced gelada genomes reveals two main populations differentiating northern (orange) and central (green) geladas. Zoo animals are of mainly central ancestry but two individuals with the highest levels of northern ancestry are also heterozygous for the centric fission characteristic of northern geladas.
Should geladas be divided into 2 species? Chiou et al. found that Northern geladas (2n = 44) have a fission of chromosome 7 compared with Central geladas (2n = 42) & all other papionins! This may affect gene flow within geladas! #2025MMM #RIP doi.org/10.1038/s415...
01.04.2025 00:35 — 👍 18 🔁 3 💬 0 📌 0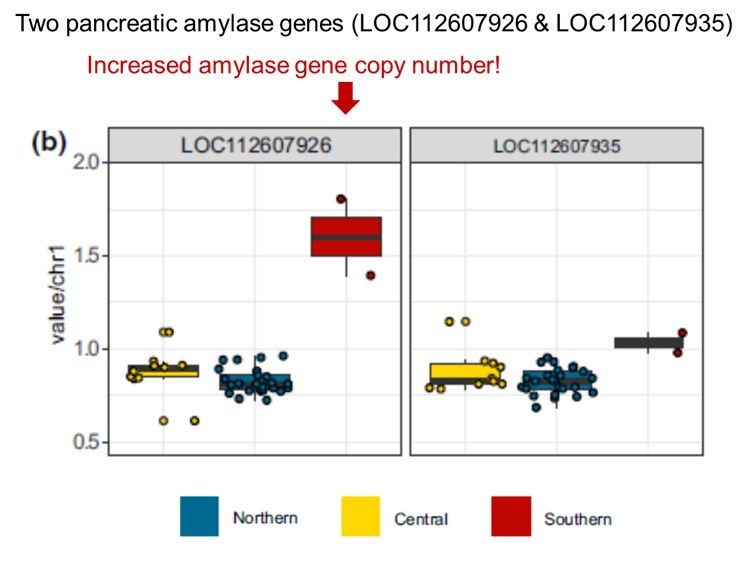
Figure 5b from Caldon et al. (2005) CNVs and amylase gene copy number. (b) Normalised genomic coverage over the two loci annotated as pancreatic amylase in geladas. Arrow added to point to the increased copy number of one (LOC112607926) of the two pancreatic amylase genes in southern geladas.
Caldon et al. (2024) sequenced genomes from the 3 major gelada pops (north, central & south) & found S pops have + pancreatic amylase gene copy #. More tuber consumption when grass is scarce may serve selection pressure for more efficient processing of starchy food! #2025MMM doi.org/10.1111/mec....
01.04.2025 00:11 — 👍 12 🔁 4 💬 0 📌 0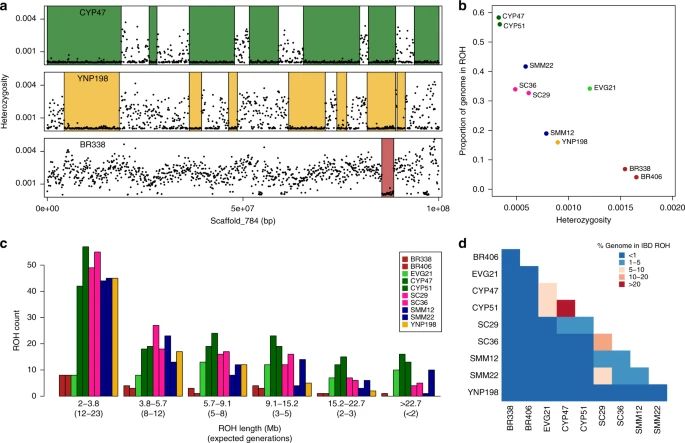
a) Sliding window heterozygosity (black dots) and called ROH (runs of homozygosity) across a single scaffold for 3 pumas from 3 different populations (Big Cypress, Yellowstone, and Brazil) b) Average genome-wide heterozygosity versus the proportion of the genome in ROH c) Distribution of lengths of ROH. d) Heat map showing the percent of the genomes that are in ROH that are shared IBD (identity by descent) between pairs of pumas.
Genome sequencing confirms that pumas colonized N. America from S. America ~100-300mya. However, fragmentation has left many N. American pops w/ low diversity and long ROH (signature of recent and severe inbreeding). #2025MMM www.nature.com/articles/s41...
01.04.2025 00:08 — 👍 21 🔁 7 💬 1 📌 1
Tonight I'll be skeeting cool genetics & evolution facts about our amazing final combatants of #2025MMM!
Skeets written by @elliecat.bsky.social @afogel29.bsky.social @acstone.bsky.social and @fervillanea.bsky.social
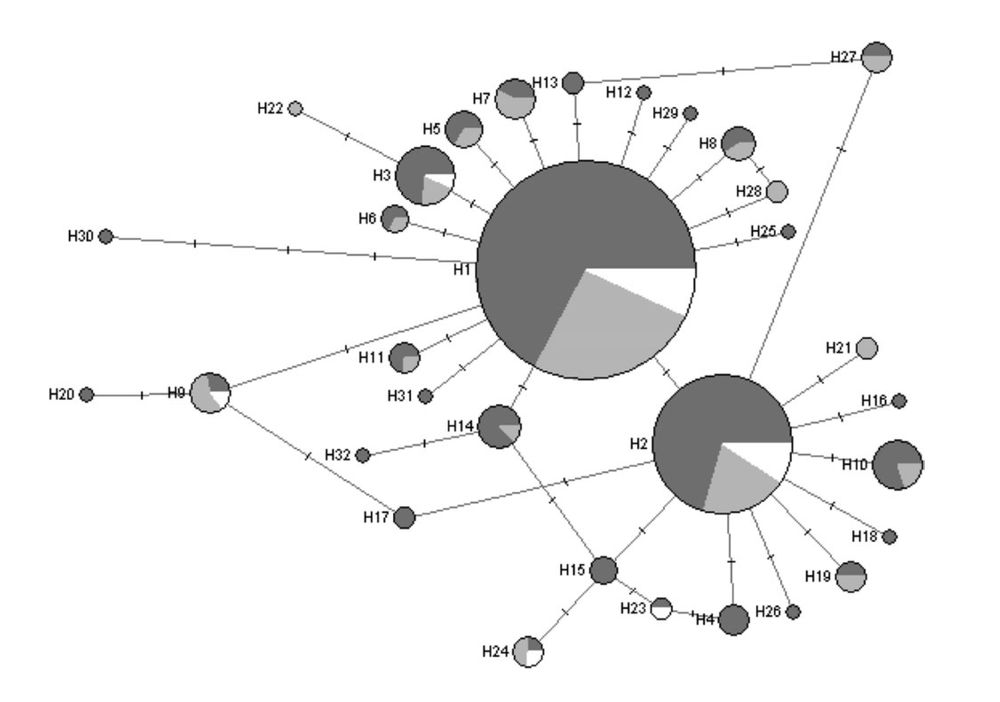
Maximum parsimony network of 32 mitochondrial control region haplotypes in American white pelicans (Pelecanus erythrorhynchos). The size of the circle is proportional to the frequency of the haplotype in the population; shading indicates the region in which the haplotype occurs (east, dark gray; west, light gray; south, white).
Great White Pelicans have high effective population sizes and gene flow, buffering against genetic erosion. Their high heterozygosity suggests that, at least in Africa, they retain robust genetic variation crucial for adaptability. #2025MMM #RIP doi.org/10.1093/jher...
14.03.2025 02:00 — 👍 14 🔁 3 💬 0 📌 0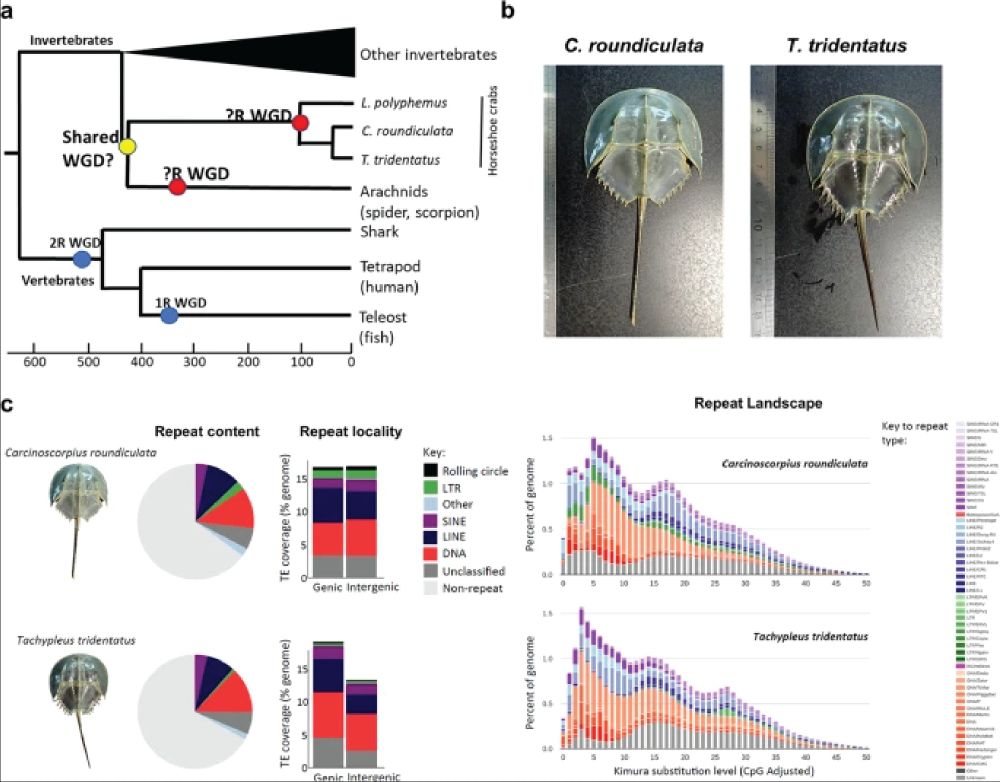
This phylogeny maps whole genome duplications across animals, hinting at shared ancestry with arachnids like spiders and scorpions. The images showcase Carcinoscorpius rotundicauda and Tachypleus tridentatus, two species with assembled genomes helping scientists decode their ancient genomic history.
Horseshoe crabs had 3 whole-genome duplications ~135 Mya, but their ties to arachnids remain unclear. Genomes of C. rotundicauda & T. tridentatus show syntenic Hox links to spiders & scorpions, fueling debate on whether they’re marine arachnids or an old sister taxa #2025MMM doi.org/10.1038/s420...
14.03.2025 01:53 — 👍 16 🔁 3 💬 0 📌 0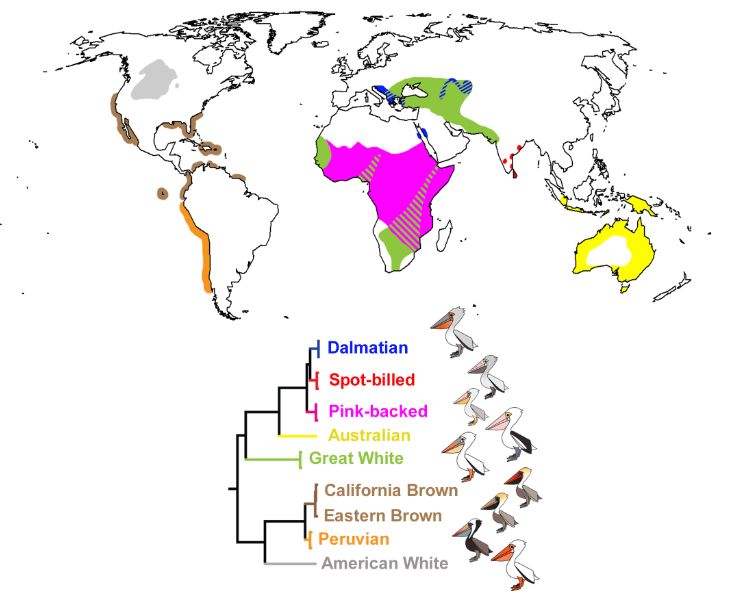
Approximate distributions of the world’s pelican species and their respective phylogenetic relationships.
Fossils show pelicans have existed since at least the early Oligocene; the oldest Pelecanus fossil is ~30 Mya, supporting deep genetic divergences among modern species. Despite spanning Africa, Europe & Asia, their DNA shows little population structure. #2025MMM doi.org/10.1016/j.ym...
14.03.2025 01:51 — 👍 14 🔁 4 💬 0 📌 0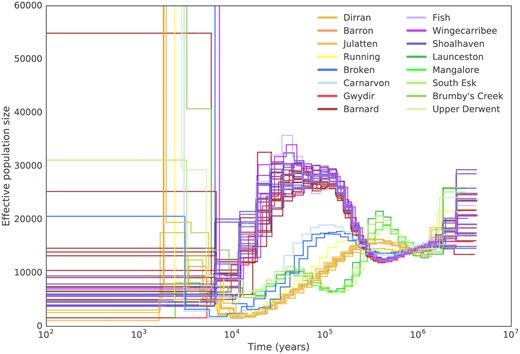
Population trajectories among four distinct Platypus population groups in Australia and Tazmania. Each group has its own trajectory, but all reach their lowest numbers of individuals at the present ime.
Martin et al 2018 whole genome data found four distinct and well-structured populations of Platypus. They find population trajectories at their lowest numbers at the most recent time. This is consistent with a decline in platypus numbers across Australia #2025MMM #RIP doi.org/10.1093/molb...
14.03.2025 01:35 — 👍 13 🔁 3 💬 0 📌 0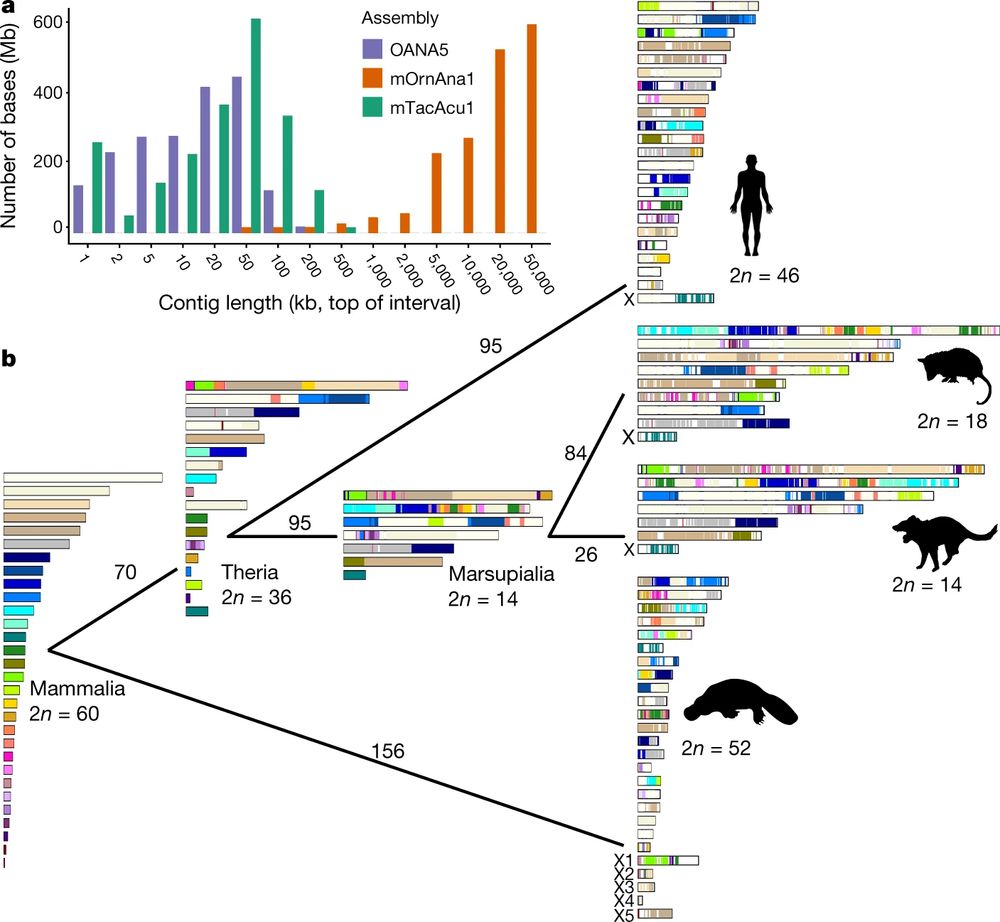
Evolution of autosomal chromosomes among mammal groups. From a reconstructed 60 chromosomes in the ancestral mammals to 52 in monotremes, 46 in humans, 14 in Tazmanian Devils and 18 in Opposums. Conversely, X chromosomes in marsupials and placental mammals share a common origin but monotremes instead present 5 unique X chromosomes.
Zhou et al 2021 sequenced the complete genome of the Platypus and Echidna and find a surprise. Monotremes have 5 pairs of sex chromosomes which have a different origin from the other mammal sex chromosomes, instead being closely related to bird autosomes doi.org/10.1038/s415... #2025MMM
14.03.2025 01:26 — 👍 21 🔁 7 💬 2 📌 0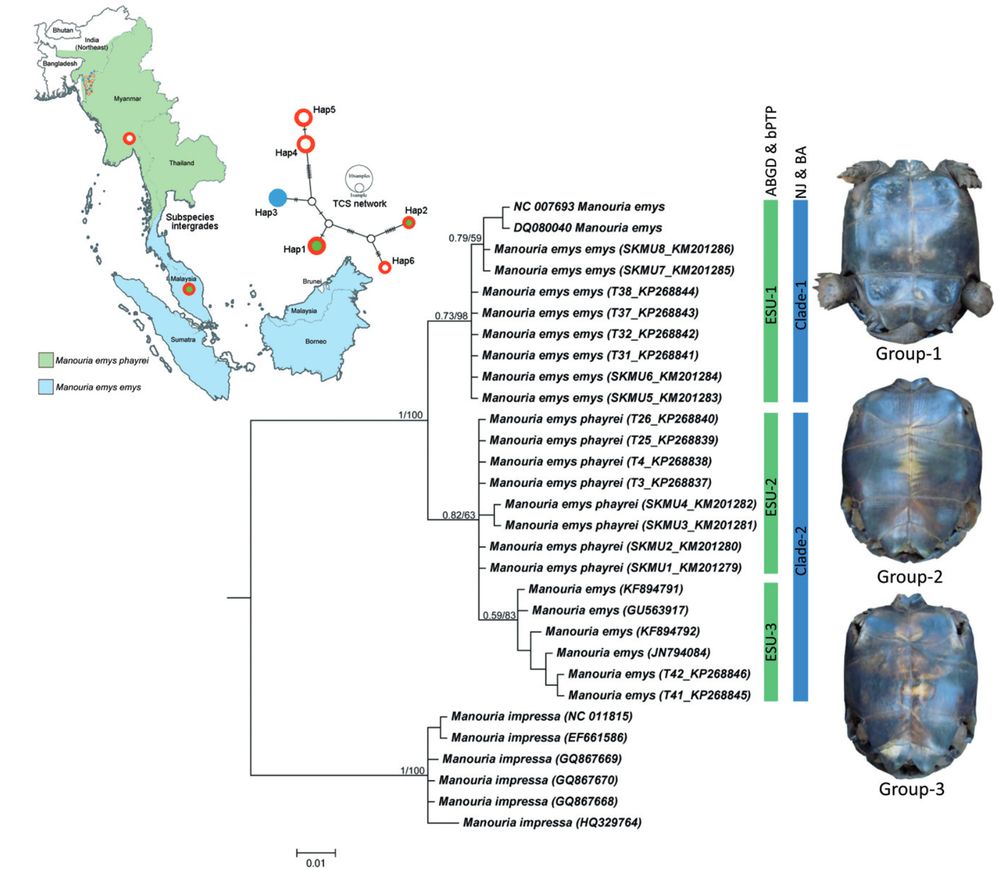
Map showing the distribution pattern of two subspecies and their intergradation zone in southern Thailand. Bayesian phylogeny on mtCOI with posterior probability and bootstrap value by NJ analysis are superimposed with each node. Plastron pattern of three morphological forms of M. emys shows with respective clades.
Plastron shape and color suggest three lineages of Forest Tortoise, but Kundu et al 2018 found that mitochondrial lineages are shallow, that is, there is no evidence of deep lineages within the species despite the morphological differences #2025MMM doi.org/10.1080/2380...
14.03.2025 01:24 — 👍 13 🔁 4 💬 0 📌 0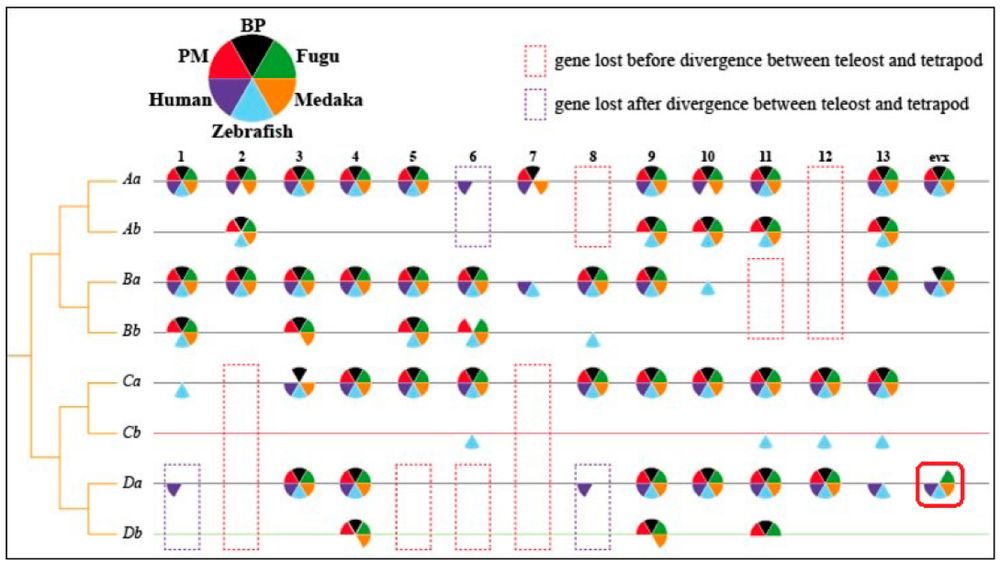
updated image from doi.org/10.3390/ani8...
14.03.2025 01:12 — 👍 0 🔁 0 💬 0 📌 0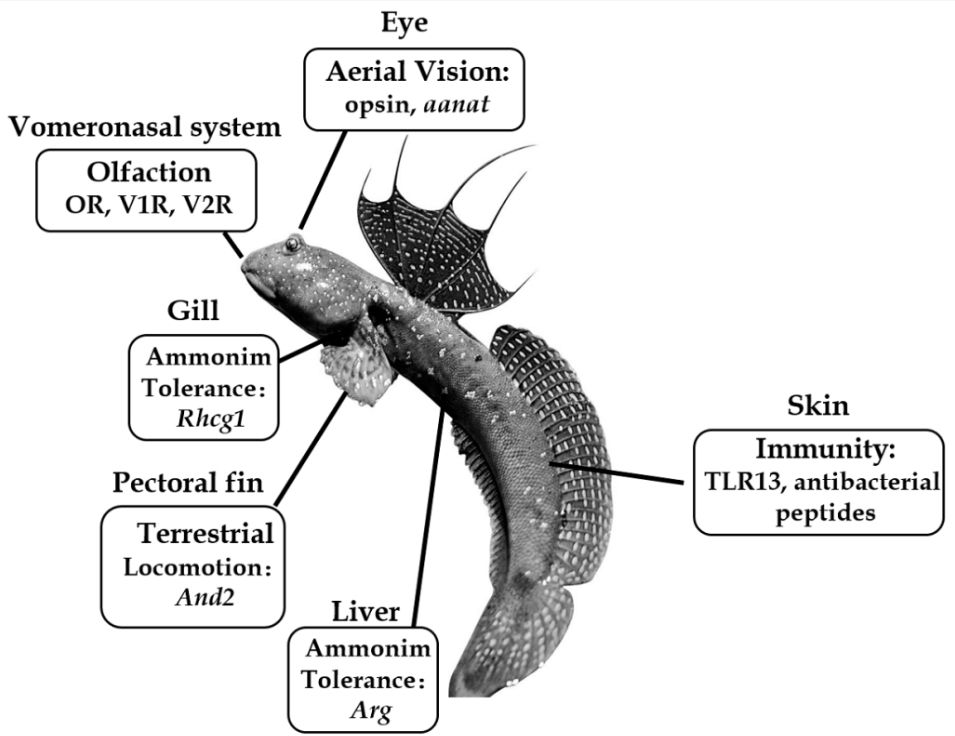
Summary of important genes related to the amphibious lifestyle of mudskippers. The blue-spotted mudskipper (B. pectinirostris, BP) is jumping on the mudflat. Abbreviations: aanat, aralkylamine N-acetyltransferase; And2, actinodin 2; Arg, arginase; Rhcg1, Rhesus C glycoprotein 1; TLR13, Toll-like receptor 13.
updated figure
14.03.2025 01:06 — 👍 0 🔁 0 💬 0 📌 0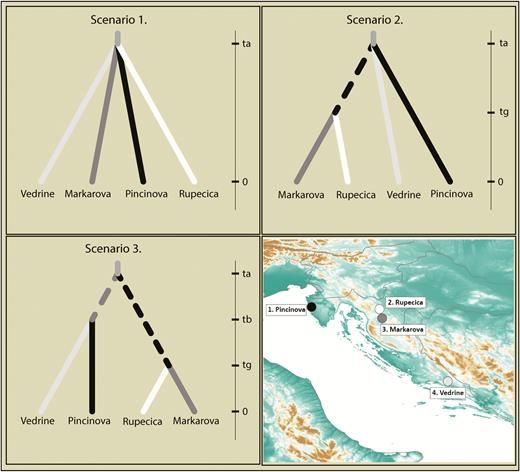
Three different scenarios for diversification of Proteus populations in Croatia tested with DIYABC 2.1. The comparison of the scenarios suggested that scenarios 2 and 3 were most likely, with scenario 3 being selected by the logistic approach (55.12% of the simulations).
Vörös et al 2019 find that genetic diversity for 4 cave populations of Olm in Croatia is low, but genetic differentiation between the 4 populations was very high, implying these might be separate species that have not admixed in millions of years #2025MMM #RIP doi.org/10.1093/jher...
14.03.2025 01:02 — 👍 16 🔁 4 💬 1 📌 0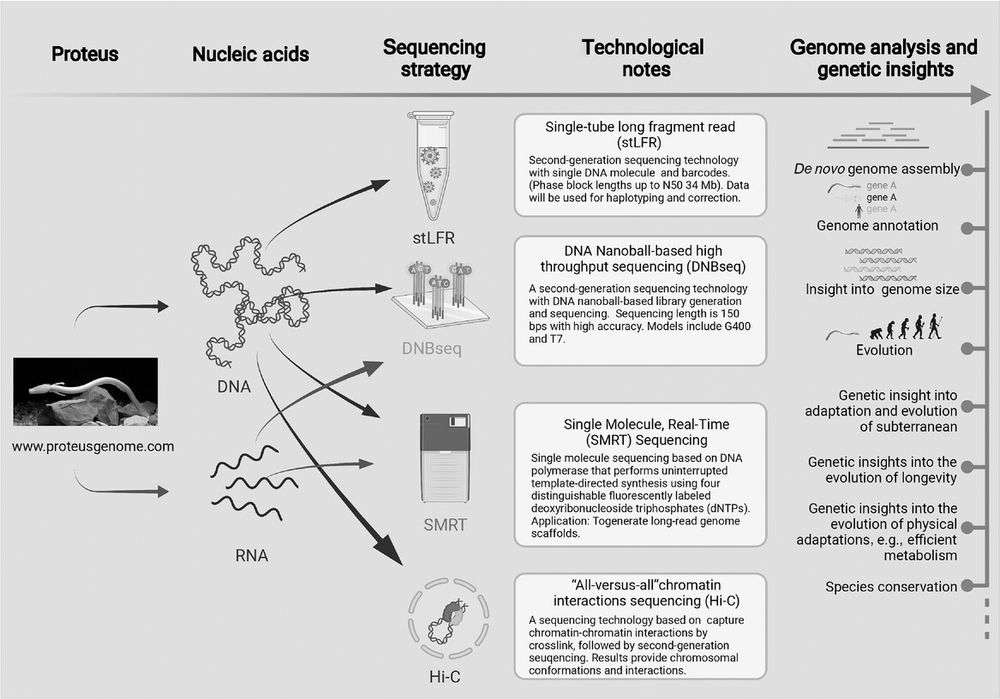
Schematic illustration of the olm genome sequencing plan. Both genomic DNA and RNA are isolated from olm tissue, followed by four sequencing strategies: stLFR, DNBseq, SMRT, and Hi-C. The proteus genome will provide genetic insights into the evolution and adaptation to the subterranean environment, longevity, metabolism, regeneration, and aid species conservation.
The Olm genome is of great interest but it remains unsequenced due to its large size. Kostanjšek et al 2021 estimate a size of ~50 Giga base pairs, comparable to the largest sequenced animal genomes so far: lungfishes and axolotl. The human genome is only 3.2 Gpb. #2025MMM doi.org/10.1111/nyas...
14.03.2025 00:52 — 👍 26 🔁 5 💬 0 📌 2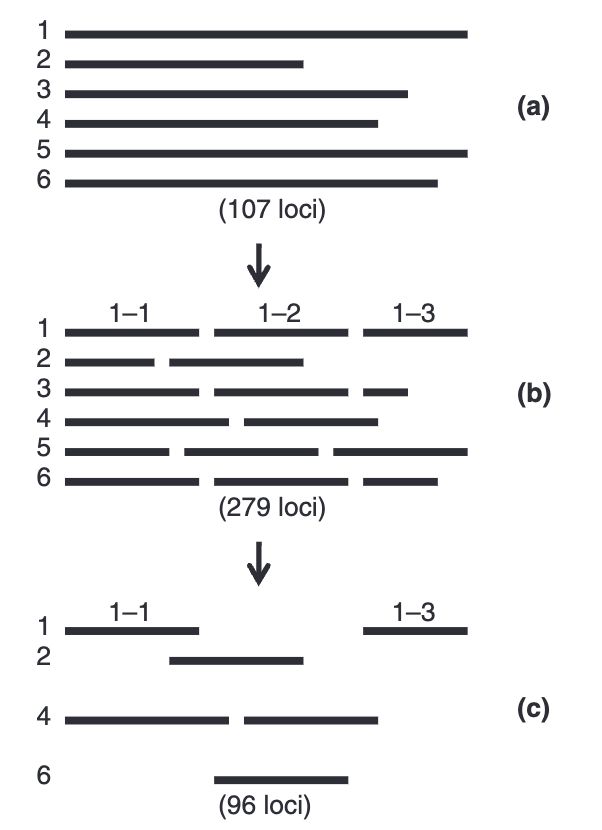
Flowchart illustrating the development and selection ofloci for re-sequencing in ringed seals. Initially, primer and refer-ence information for 107 loci varying in length from a few 100 tomore than 1000 bp was supplied by us to polymorphic, as repre-sented by locus 1–6 in (a). These 107 loci were split up in 279shorter loci of 200–300 bp in length, as illustrated in (b). Follow-ing primer design, optimization and quality control, these 279loci were reduced to 96 loci, as illustrated in (c). Note that amongthese final 96 loci, some are located on the same initial locus andare thus known to be physically linked, e.g. 1-1 and 1-3 in (c).
Assembling a new genome is difficult, normally we use the genome of a close species as a template, but relicts don't have close species. Olsen et al 2011 find a workaround to get some genetic data for the ringed seal until we have a complete genome #2025MMM doi.org/10.1111/j.17...
14.03.2025 00:51 — 👍 9 🔁 2 💬 0 📌 0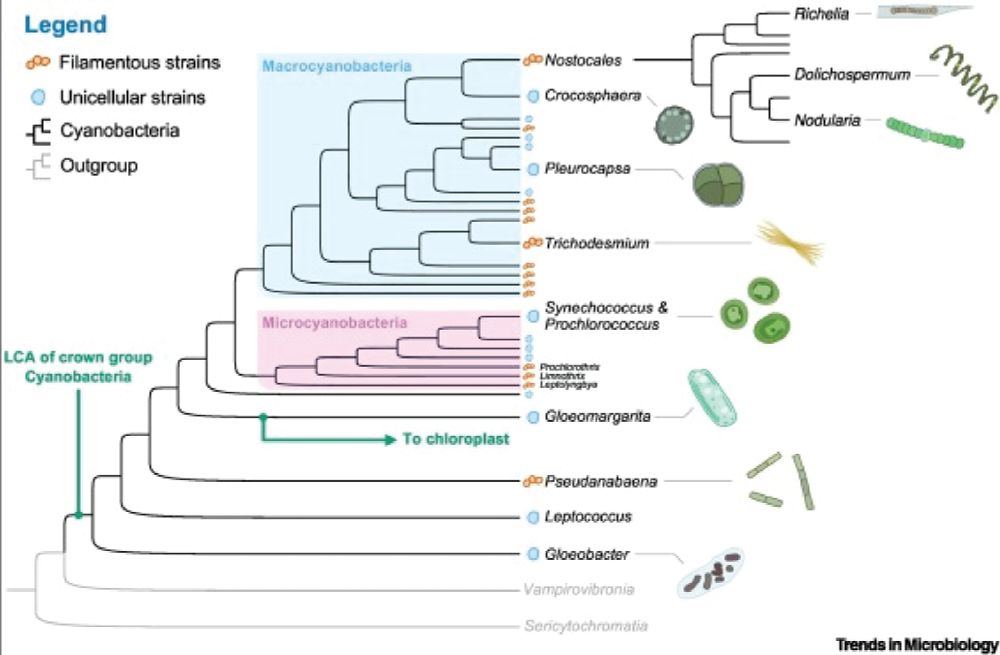
A cladogram traces cyanobacterial evolution, linking them to nonphotosynthetic relatives, Vampirovibrionia and Sericytochromatia. Phylogenetic analysis places the last common ancestor of crown Cyanobacteria ~2.5 Ga, highlighting key transitions, including the lineage that gave rise to chloroplasts.
The core proteins of PSII predated the Great Oxygenation Event (~2.3 Ga). Most cyanobacterial lineages, including chloroplast ancestors, diversified later, with planktonic cyanobacteria rising to dominance by the Precambrian’s end, shaping Earth's carbon cycle #2025MMM #RIP 10.1016/j.tim.2021.05.008
14.03.2025 00:26 — 👍 18 🔁 3 💬 2 📌 0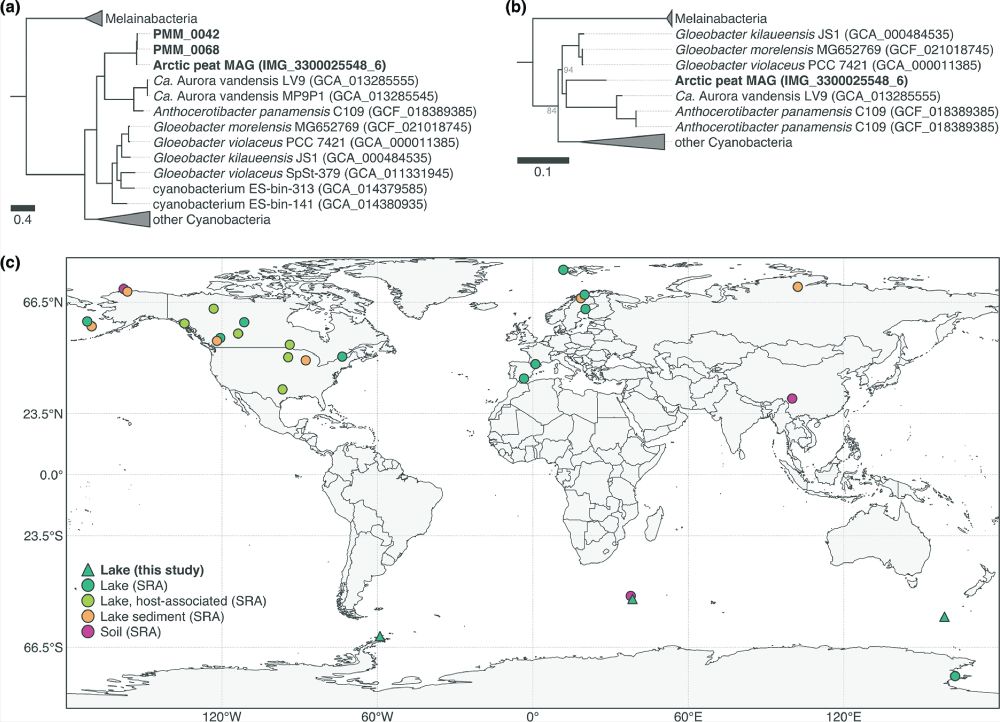
A newly identified early-branching Cyanobacterium, Ca. Sivonenia alaskensis, thrives in cold environments worldwide. Phylogenetic trees place it within Gloeobacterales, while global metagenomic data reveal its widespread presence in polar and alpine lakes, reshaping our understanding of cyanobacterial evolution.
Polar lakes are dominated by Cyanobacteria, yet few polar genomes have been sequenced. Using genome-resolved metagenomics, 37 MAGs were recovered from Arctic, sub-Antarctic, and Antarctic mats, revealing 17 species, including a novel cold-adapted lineage. #2025MMM doi.org/10.1099/mgen...
14.03.2025 00:20 — 👍 15 🔁 2 💬 0 📌 0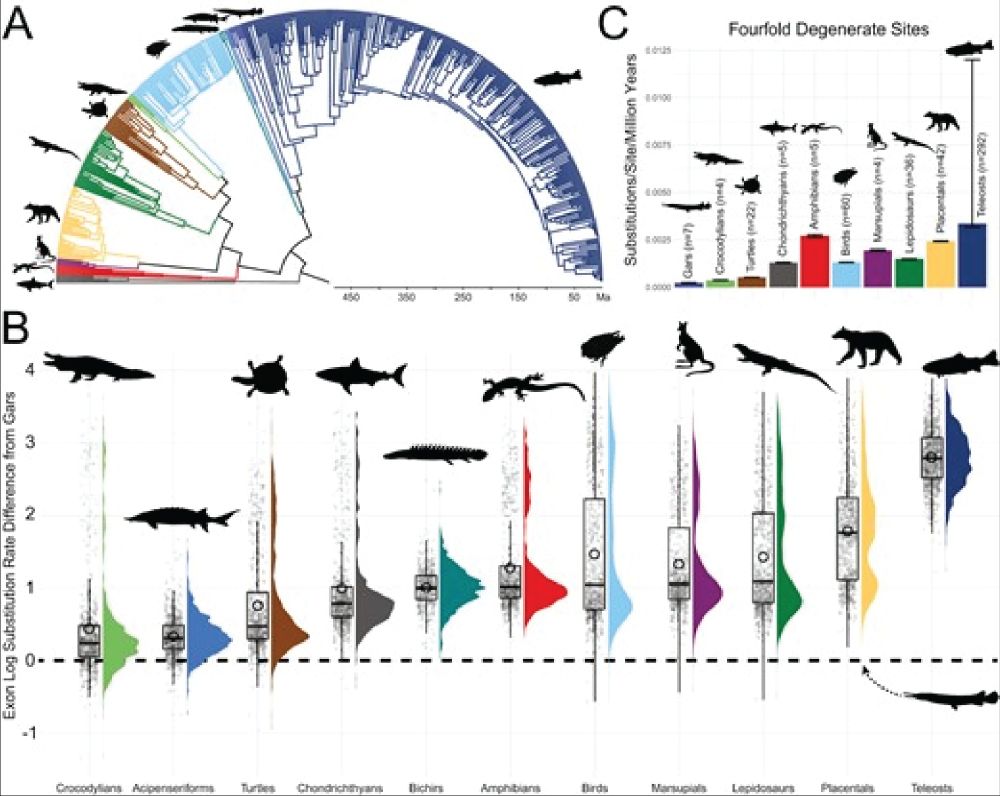
Genomic substitution rates across vertebrates highlight the slow molecular evolution of gars. A time-calibrated phylogeny of 478 vertebrates shows divergence dates. Violin and box plots display exon log substitution rates across clades, and substitution rates at fourfold degenerate sites are compared.
Evolutionary stasis occurs when lineages show little phenotypic change. Gars were found to have one of the lowest substitution rates among jawed vertebrates. Two gar species, diverged for over 100 my, were shown to hybridize—the oldest known case in eukaryotes #2025MMM doi.org/10.1093/evol...
14.03.2025 00:19 — 👍 44 🔁 10 💬 3 📌 2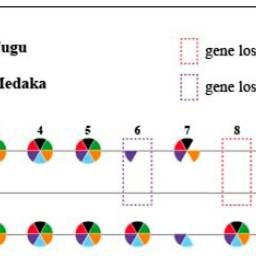
Hox gene clusters in fish and human. The red solid box highlighted the loss of hoxDa-evx of Boleophthalmus pectinirostris and Periophthalmus magnuspinnatus, which may generate neurological and morphological effects during development. Numbers 1–13 and evx are the general codes for hox genes.
Mudskippers famously “walk” on land using their front fins. Genome scans identified changes in certain Hox genes and show unique indels in a limb-related transcription factor gene (Tbx2) as well as modifications in genes involved in fin ray formation. #2025MMM #RIP
14.03.2025 00:13 — 👍 19 🔁 5 💬 2 📌 2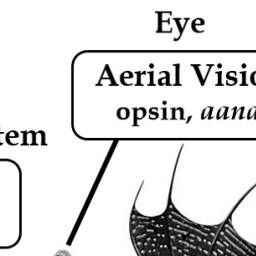
Summary of important genes related to the amphibious lifestyle of mudskippers. The blue-spotted mudskipper (B. pectinirostris, BP) is jumping on the mudflat. Abbreviations: aanat, aralkylamine N-acetyltransferase; And2, actinodin 2; Arg, arginase; Rhcg1, Rhesus C glycoprotein 1; TLR13, Toll-like receptor 13.
Mudskippers have evolved visual adaptations for seeing in air. Comparative genomics showed that certain vision-related genes, like aanat1a, LWS1 and LWS2, are lost or altered in P. schlosseri, aligning with its need for aerial vision #2025MMM doi.org/10.3390/ani8...
14.03.2025 00:07 — 👍 14 🔁 3 💬 1 📌 0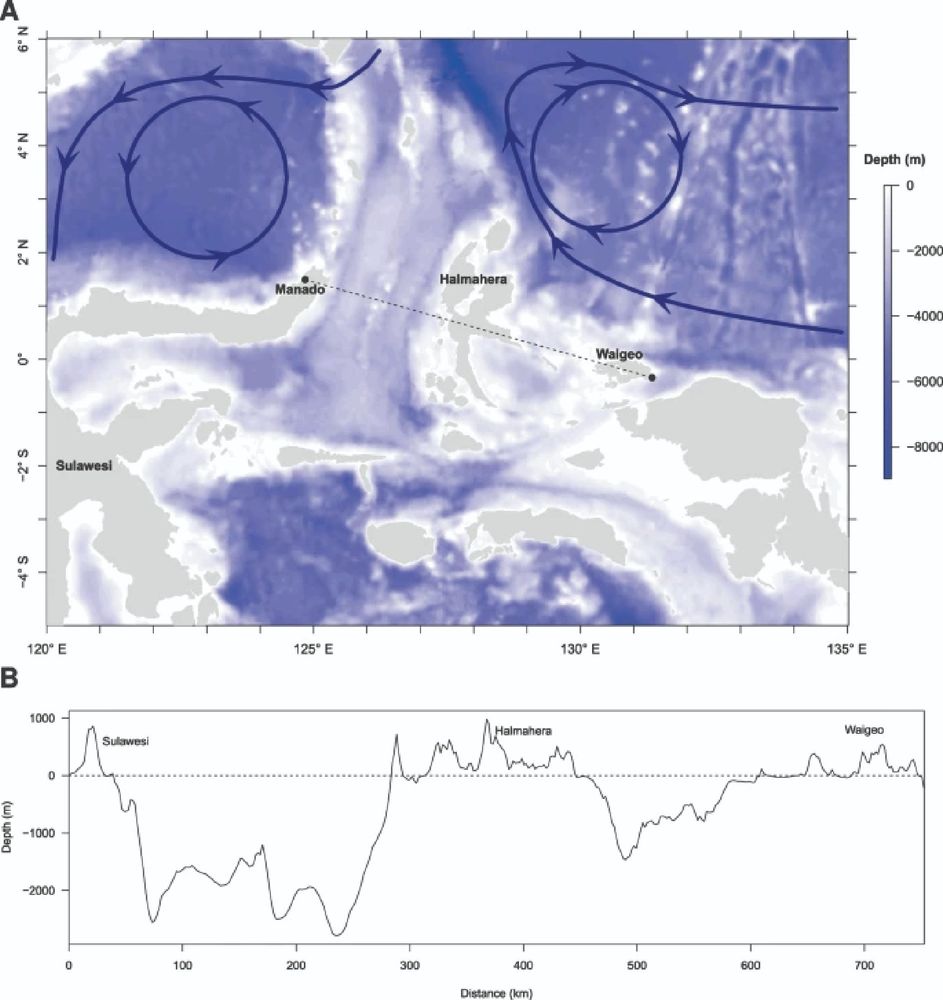
Bathymetry of Eastern Indonesia depicting the oceanic currents and oceanic depth between two populations of Latimeria menadoensis. The arrows show the main surface currents. (B) Bathymetry and elevation profile between Manado and Waigeo.
Despite being members of the same species, two populations of Coelocanths in eastern Indonesia (Latimeria menadoensis) were 13 million years diverged. Isolation of these two populations is likely biogeographical in response to oceanic currents. #2025MMM doi.org/10.1038/s415...
14.03.2025 00:06 — 👍 24 🔁 3 💬 1 📌 2Tonight I'll be posting fun genetic facts about our awesome competitors in #2025MMM
13.03.2025 23:47 — 👍 9 🔁 1 💬 0 📌 0People keep saying "I can easily tell when students use LLMs". People are wrong.
06.01.2025 14:07 — 👍 13 🔁 6 💬 0 📌 0AI's ability to have personalized and dialogue based conversation might be more effective than other methods to counter misinformation. Very exciting work and related to the ideas being tested by @ismaharif.bsky.social at @mediamosaiclab.bsky.social
27.12.2024 19:17 — 👍 8 🔁 3 💬 0 📌 1