To all the #RNAFramework users who updated to v2.9.6: please issue a "git pull" to ensure the latest Core::Utils module gets updated as this escaped the last commit. Apologies for the inconvenience!
11.02.2026 11:31 —
👍 1
🔁 0
💬 0
📌 0

Beyond significant speed-ups, this version features a complete rewrite of the rf-correlate module for calculating pairwise correlations between samples. The new version has no sample number limit, and it generates clustered correlation heatmaps.
09.02.2026 10:52 —
👍 2
🔁 0
💬 0
📌 0
#RNA Framework 2.9.6 is out! Several additions, important fixes and improvements. Please update! Check it out: buff.ly/4guLWsP & see the CHANGELOG (buff.ly/4gtJ67u) for the details!
09.02.2026 10:49 —
👍 4
🔁 0
💬 1
📌 0
draco - DRACO
Second: DRACO v1.3 is out! Greatly improved algorithm for increased sensitivity and accuracy in the reconstruction of #RNA conformation reactivity profiles. The biggest improvement: replicate support (for more information, check the documentation: shorturl.at/zY3nu). Check it out: lnkd.in/gBhpnisG
12.12.2025 19:23 —
👍 2
🔁 1
💬 0
📌 0
The biggest improvement is the introduction of paired-end support in rf-count (for more information, check the documentation: shorturl.at/LZcou). (2/n)
12.12.2025 19:23 —
👍 1
🔁 0
💬 1
📌 0
As it's almost Christmas, here're 2 presents from the lab! 🎄
First: #RNA Framework 2.9.5 is out! Several additions, fixes and improvements. Check it out: lnkd.in/gi3QDNeq & see the CHANGELOG (shorturl.at/Lcobs) for the details! (1/n)
12.12.2025 19:23 —
👍 2
🔁 1
💬 1
📌 0
Until a fix will be released, we advice users to install ViennaRNA v2.7.0 in a separate folder and to provide the path to RNAplot v2.7.0 via the -vrp parameter of rf-fold. (4/n)
13.10.2025 12:47 —
👍 0
🔁 0
💬 0
📌 0
!!! Important note: A bug was introduced in ViennaRNA v2.7.0, which breaks the pseudoknot detection functionality of rf-fold. However, RNAplot v2.7.0 is required to take advantage of the new secondary structure plotting functionality. (3/n)
13.10.2025 12:47 —
👍 0
🔁 0
💬 1
📌 0

Highlights: new normalization method in rf-norm (Mitchell method, pubmed.ncbi.nlm.nih.gov/37334863/), statistics plots in rf-count & rf-count-genome, and secondary structure plots in rf-fold (powered by ViennaRNA v2.7.0) (2/n)
13.10.2025 12:47 —
👍 0
🔁 0
💬 1
📌 0

Please join us for our next seminar, featuring Danny IncaRNAto, PhD: “Discovery of RNA regulatory structural switches via ensemble mapping”
@incarnatolab.bsky.social #rna #casprnasig #rnastructure
10.10.2025 17:34 —
👍 7
🔁 2
💬 0
📌 1
Looking forward!
10.10.2025 18:20 —
👍 1
🔁 0
💬 0
📌 0

Two group leader positions available in the broader areas of RNA science, RNA technologies, and RNA medicine. Attractive packages and a great environment. Come and join us at Helmholtz RNA Würzburg, Bavaria.
08.10.2025 21:45 —
👍 68
🔁 80
💬 1
📌 1
Check out this new fantastic method from @rivaselenarivas.bsky.social to predict #RNA structure + 3D motifs and pseudoknots guided by covariation
06.10.2025 11:46 —
👍 3
🔁 0
💬 0
📌 0
Happy #RNA day to all my fellow RNA lovers!
01.08.2025 10:05 —
👍 4
🔁 0
💬 0
📌 0
Thanks Lars! It definitely was :-)
30.07.2025 08:47 —
👍 1
🔁 0
💬 0
📌 0
Thanks Dirk-Jan!
29.07.2025 16:54 —
👍 1
🔁 0
💬 0
📌 0
Thanks Rita!
29.07.2025 16:54 —
👍 1
🔁 0
💬 0
📌 0
Thanks Fede!
28.07.2025 12:07 —
👍 1
🔁 0
💬 0
📌 0
Thanks Sebastian!
27.07.2025 20:11 —
👍 0
🔁 0
💬 0
📌 0
Thanks Ivano!
27.07.2025 17:25 —
👍 1
🔁 0
💬 0
📌 0
And a huge thanks to @erc.europa.eu for funding that made all of this possible! (11/n)
27.07.2025 17:03 —
👍 3
🔁 0
💬 0
📌 0
Shoutout to rockstar Ivana Borovská, who led this incredible work, and to our collaborators, particularly @mtwolfinger.bsky.social and Wim Velema! (10/n)
27.07.2025 17:03 —
👍 4
🔁 1
💬 1
📌 0
While our study still presents many limitations, we sincerely hope this will represent a stepping stone in better understanding and characterizing the structural dynamics of cellular RNAs, and their roles in regulating gene expression. (9/n)
27.07.2025 17:03 —
👍 6
🔁 1
💬 1
📌 0
papers/Borovska et al., 2025 at main · dincarnato/papers
Data & pipelines associated with papers from the lab - dincarnato/papers
Furthermore, we introduced the DeConStruct framework (github.com/dincarnato/p...), to aid the discovery and prioritization of conserved regulatory RNA structural switches discovered via ensemble deconvolution analysis of chemical probing data. (8/n)
27.07.2025 17:03 —
👍 3
🔁 1
💬 1
📌 0
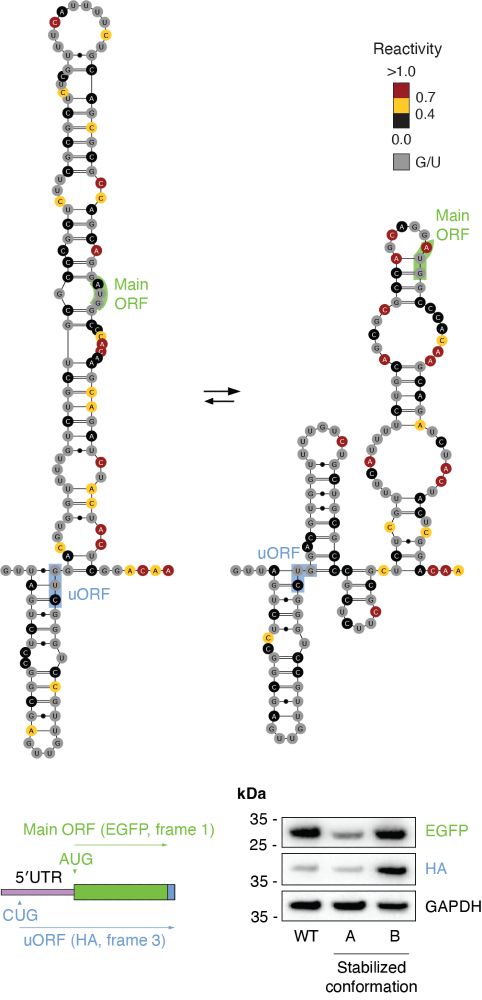
For example, the 5′-UTR of the CKS2 RNA populates two conformations, one promoting increased translation of a uORF, and one reducing translation of the main ORF. (7/n)
27.07.2025 17:03 —
👍 3
🔁 1
💬 1
📌 0
Thanks to this method, we observed that regions of human 5′-UTRs populating 2+ alternative conformations, tend to be enriched for upstream start codons (NTG), suggesting that the switch between these conformations might regulate translation of uORFs vs. main ORF. (6/n)
27.07.2025 17:03 —
👍 4
🔁 1
💬 1
📌 0
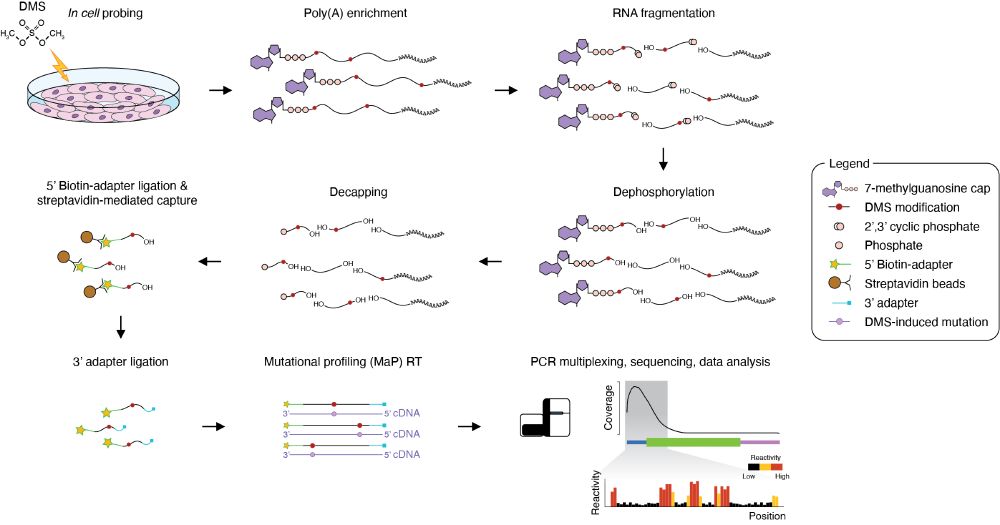
Next, we sought to explore the transcriptome of human cells. Due to the extreme sequencing depth required for ensemble deconvolution analyses, we developed 5′UTR-MaP, which enables the transcriptome-scale interrogation of 5′-UTRs (and, in general, of 5′-terminal regions). (5/n)
27.07.2025 17:03 —
👍 4
🔁 1
💬 1
📌 0
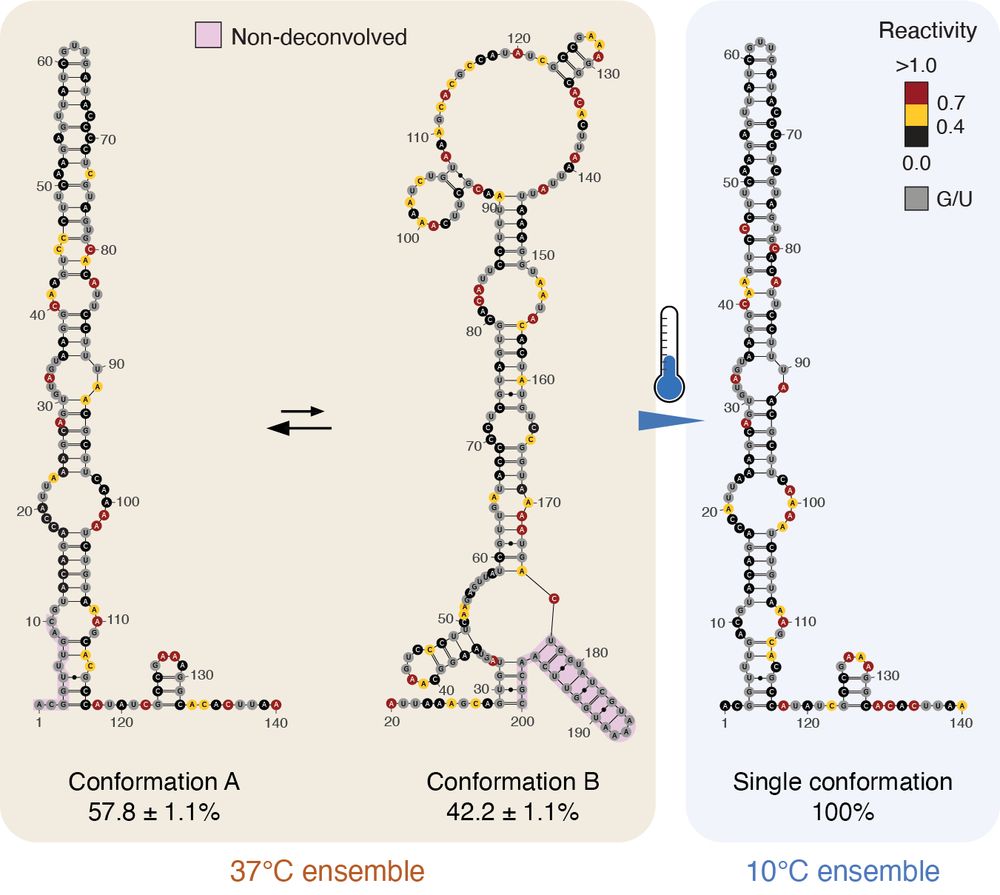
Interestingly, the well characterized cspA thermometer already populates both ON and OFF conformations at 37°C, and completely shifts to the ON conformation at 10°C, explaining why cspA is among the top-expressed proteins in the E. coli proteome. (4/n)
27.07.2025 17:03 —
👍 5
🔁 0
💬 1
📌 0









