
@inmice.bsky.social
Here's one for you
www.sciencedaily.com/releases/202...
@ericfauman.bsky.social
GWAS whisperer. It's usually the closest gene. Executive Director, Integrative Biology, Internal Medicine Research Unit. Pfizer R&D. All views my own. Also cats.

@inmice.bsky.social
Here's one for you
www.sciencedaily.com/releases/202...

If you didn't get your certificate were you really even there?
19.10.2025 17:58 — 👍 5 🔁 0 💬 0 📌 0I did filter to p < 5e-8. But we also some filtering to independent lead snps. We also have sissies studies ranging from n of a few hundred to a few million subjects.
28.09.2025 15:14 — 👍 1 🔁 0 💬 0 📌 0
It's a little odd. I am plotting the log10 of the -log10(pvalue) on the x axis so in this plot the null signal drops off much faster than what we observe in the @gwascatalog.bsky.social
I'm curious if anyone has a hypothesis why the lead SNP p-values would have this distribution.
Sounds great!
28.09.2025 11:12 — 👍 0 🔁 0 💬 0 📌 0The effect size in gwas catalog are harder to compare because they are not necessarily in standard deviation units.
28.09.2025 01:07 — 👍 0 🔁 0 💬 1 📌 0Oh you win! Yes, all are inappropriate attempts to avoid underflows.
28.09.2025 01:05 — 👍 1 🔁 0 💬 1 📌 0Bonus question. Why is the plot of log10(count) as a function of log10(-log10(p)) a straight line?
Recall, the data is all the lead SNPs recorded in the GWAS catalog.

Quick quiz. This is a plot of how often each p-value appears in this @gwascatalog.bsky.social. Actually I binned the -log10(p) values to their integer values (e.g., 8-9, 9-10, etc).
I'm highlighting 3 outliers here, in orange.
What are these p-values and why do they diverge so much?

Getting ready for ASHG25 here in Boston! Who's coming to town?
As you know, I am fascinated by extreme GWAS p-values.
Will we see anything beyond the current record holder, 10^-28538, for eye color.
Interestingly, the p-values for all lead SNPs in the @gwascatalog.bsky.social follow a power law
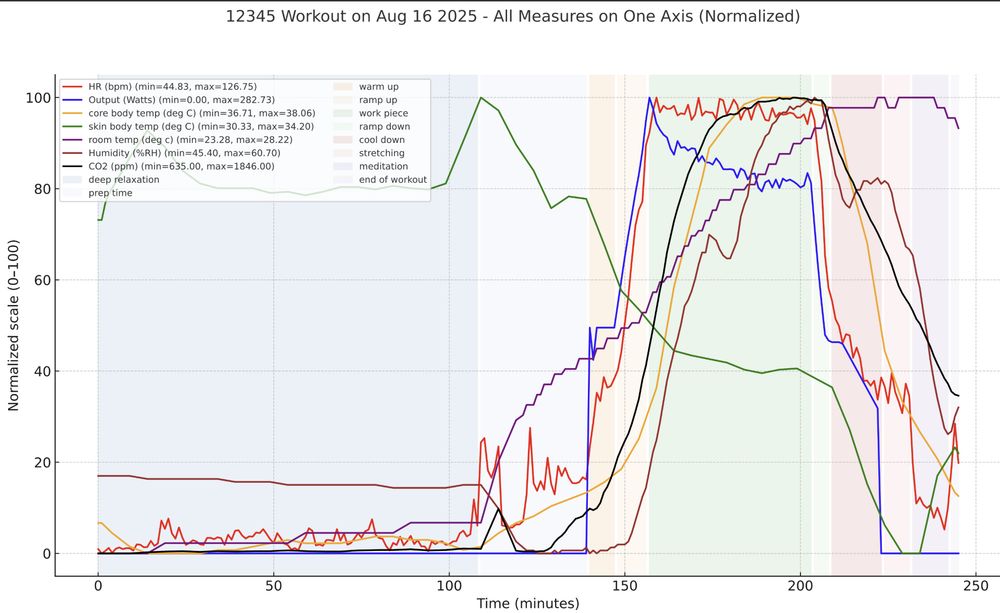
For fun I'm also tracking the room temperature, humidity and CO2 which all increase during the workout, as expected.
All the analysis, charting and graphs were done by GPT5 operating on a google sheet where I collected the minute by minute stats.

GPT4o pointed me to a device that measures core and skin body temperature. In gold you can see my core body temp continues to rise even after I reach my max heart rate (red), thus requiring further decreases in the work output (blue). Skin temperature drops during the workout, thanks to fans + sweat
16.08.2025 22:49 — 👍 0 🔁 0 💬 1 📌 0
This graph plots a study I did on myself which involves trying to maintain a heart rate of exactly 123 bpm for 45 minutes. Doing so requires adjusting the work output. In this plot the red line is my heart rate while the blue line is the work output, which you can see decreases over time.
16.08.2025 22:46 — 👍 0 🔁 0 💬 2 📌 0
Specifically, the phenomenon is called cardiovascular drift and it is a consequence of the body having to do the mechanical work and then also additional work to dissipate the heat generated by the effort, especially the vasodilation to direct internal body heat to the surface.
16.08.2025 22:44 — 👍 1 🔁 0 💬 1 📌 0
Years ago I asked on Twitter why my heart rate would continue to climb even after I fixed the speed and incline on my treadmill. At the time, no one on Twitter could give me a satisfactory answer.
But now GPT4o was able to answer it and suggested approaches to quantify the phenomenon.
Isn't that the beginning of the movie "Brazil"?
16.08.2025 22:37 — 👍 2 🔁 0 💬 1 📌 0This was the random page on the internet that set me down this rabbit hole:
www.tomsguide.com/wellness/fit...
DeGETTE: Will you commit that neither you or anyone else at HHS will take retaliatory action against the brave scientists at NIH? Should be an easy answer bc it's illegal
RFK Jr: I can commit that we are absolutely depoliticizing science at NIH for the first time
D: I asked you a simple question
It could be like designing an antibody to reduce circulating cystatin C levels in an attempt to improve kidney function...
23.06.2025 03:57 — 👍 3 🔁 1 💬 1 📌 0And if that's the case, then we haven't really established a causal link between making people walk faster and seeing them age slower.
23.06.2025 03:30 — 👍 2 🔁 0 💬 1 📌 0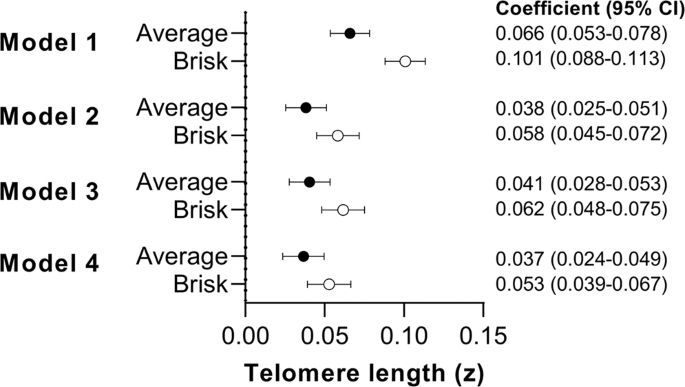
This makes no sense to me.
How does genetically predicted walking speed relate to biological aging (here as telomere length( except as some sort of biomarker of overall health?
www.nature.com/articles/s42...

To add to your weekend reading, check out this nice report on the Genes & Health cohort from Hye In Kim, David van Heel and many many more:
www.medrxiv.org/content/10.1...
Signed
26.05.2025 22:40 — 👍 3 🔁 0 💬 0 📌 0Please consider signing.
26.05.2025 22:40 — 👍 3 🔁 0 💬 0 📌 0Thanks for these caveats.
I guess I'm just surprised a major chromosomal rearrangement can fix in a population so quickly with no apparent impact on the effective population size. Is there good literature on this across other species.
(I did hedge a bit in my opening post calling it a "possible bottleneck")...
25.05.2025 21:28 — 👍 1 🔁 0 💬 0 📌 0Pirruccello #eshg2025 - “There is really one game in town right now that is @ukb UKB.” - no one else is producing image data in healthy heart at this scale. @ukbiobank.bsky.social Taking a deep-dive into image-derived phenotypes such as sphericity and cardiac age acceleration.
25.05.2025 13:18 — 👍 7 🔁 2 💬 1 📌 0This looks great. Reminds me to set aside time to read the whole thing.
25.05.2025 21:12 — 👍 2 🔁 0 💬 1 📌 0Yes, I saw that paper but don't know enough to weigh the claims of either paper. Have you registered an opinion?
We do know chimps have 48 chromosomes and we have 46 so there must have been a first fusion carrier and that must have caused some reproductive difficulties.
Interestingly the original paper doesn't say the 99% went extinct, it says they were "lost" to our lineage, which is consistent with my model.
The quote: "98.7% of human ancestors were lost at the beginning of the bottleneck, thus threatening our ancestors with extinction".