
GBE | De Novo Gene Emergence: Summary, Classification, and Challenges of Current Methods
Grandchamp, @drdomain.bsky.social et al. publish a new Review on commonly used methods for de novo gene detection, address the limitations of nomenclature and detection methods, and establish a de novo gene annotation format to standardize reporting
🔗 doi.org/10.1093/gbe/evaf197
#genome #evolution
19.11.2025 11:52 — 👍 14 🔁 6 💬 1 📌 1
PSF Fundraiser 2025
The Python Software Foundation is the charitable organization behind the Python programming language.
The PSF's 2025 end-of-year fundraiser is live 🐍🚀
#Python is for everyone—and it takes everyone to keep it thriving. Support the PSF, the Python community, and the language we love.
Join in today 💛💙 donate.python.org
#PythonForEveryone
12.11.2025 17:06 — 👍 32 🔁 24 💬 1 📌 3
YouTube video by ERGA - European Reference Genome Atlas
What is ERGA? - European Reference Genome Atlas
Sign up today to receive the monthly newsletter from the ERGA European Reference Genome Atlas to learn about #biodiversy #genomics initiatives in #Europe, courses, jobs, conferences, & more www.youtube.com/watch?v=kYL9...
27.09.2025 14:27 — 👍 10 🔁 5 💬 0 📌 0
Validate User
check out the accompanying paper doi.org/10.1093/gbe/... as well!
12.11.2025 15:45 — 👍 1 🔁 1 💬 0 📌 0
Validate User
check out the accompanying paper doi.org/10.1093/bioi... as well!
12.11.2025 15:44 — 👍 1 🔁 0 💬 0 📌 0
Validate User
Grateful to my co-authors @anna-grandchamp.bsky.social , @margaubel.bsky.social , @lacholt.bsky.social, Victor Luria and Amir Karger — great teamwork all around! 🙌
Let’s keep the conversation going — what questions does this raise for your own research? 🔍
🧬 Read more: doi.org/10.1093/gbe/...
12.11.2025 15:42 — 👍 1 🔁 0 💬 1 📌 0

🧩 Thrilled to share our new paper in @genomebiolevol.bsky.social !
We present a comprehensive review of de novo gene emergence — providing a classification of current detection methods and a roadmap for addressing major challenges in the field of gene birth from non-genic sequences.
12.11.2025 15:42 — 👍 10 🔁 6 💬 2 📌 0
Validate User
Big thanks to @anna-grandchamp.bsky.social , @margaubel.bsky.social , @lacholt.bsky.social and many others for making this happen!
We’d love to hear from the community — how could this be most useful in your workflows? 💬
📄 Read it: doi.org/10.1093/bioi...
💻Try it: github.com/EDohmen/denofo
12.11.2025 12:41 — 👍 0 🔁 0 💬 1 📌 0

🚀We’re excited to share our new paper in Bioinformatics!
We introduce a user-friendly toolkit that implements our novel DeNoFo file-format for standardised annotation of de novo gene detection workflows — enabling reproducible methodology descriptions and easier dataset comparison across studies.
12.11.2025 12:41 — 👍 19 🔁 8 💬 2 📌 0
PhD position available in evolutionary genomics/bioinformatics (hoehnalab.github.io/job_adverts/...). Topic: analyzing gene expression evolution across several firefly species and linking expression changes to genomic architecture. The position is jointly supervised with @anaevolcatalan.bsky.social
11.11.2025 09:00 — 👍 45 🔁 54 💬 4 📌 2
The DeNoFo format & toolkit for annotating, assessing and comparing the tools and tresholds used in studies of de novo evolved genes is out now in Bioinformatics! Great effort led by @drdomain.bsky.social and Anna Grandchamp!
academic.oup.com/bioinformati...
03.11.2025 19:47 — 👍 12 🔁 6 💬 1 📌 0

BIG ANNOUNCEMENT📣: I haven’t been this excited to be part of something new in 15 years… Thrilled to reveal the passion project I’ve been working on for the past year and a half!🙀🥳 (thread 👇)
15.10.2025 12:22 — 👍 492 🔁 185 💬 56 📌 61
I’ll be advertising a post-doc position (up to 3 year) soon. It will be for somebody that is a good programmer, interested in evolution & is keen to learn new machine learning & AI approaches. I don’t have a link to a job advertisement yet. The post will be hybrid working and based at U of Liverpool
09.09.2025 14:25 — 👍 4 🔁 3 💬 0 📌 0
Going into my last week @uni-muenster.de and was really happy to have a final meeting with so many awesome people from @gevol.bsky.social . Good luck to all of you for your second phase projects, a great time and hopefully I'm crossing paths with some of you in the future! ☺️
08.09.2025 08:39 — 👍 6 🔁 0 💬 0 📌 0

Only 10 days left to apply:
We are searching for a senior postdoc (3 +3 years) in the field of theoretical ecology and evolution.
The position provides the opportunity to closely interact with experimentalists and develop own research projects.
Please RT.
Details 👇:
shorturl.at/iiiOv
01.09.2025 05:54 — 👍 26 🔁 43 💬 1 📌 3
Robert Waterhouse at #ESEB2025 - PIs/Profs please give your students a break when they're doing orthology, ancestral state reconstruction, gene/species tree reconciliation work - it's not an overnight process, there are no push-button tools, and fast doesn't equal correct! Love this :-)
19.08.2025 12:10 — 👍 21 🔁 5 💬 0 📌 0
116 FB 5 Research Assistant (m/f/d) field of Theoretical Ecology and Evolution or Computational Biology: Uni Osnabrück
Me and my group are searching for a senior postdoc (3 +3 years) in the field of theoretical/ computational biology.
The position provides the opportunity to closely interact with experimentalists and develop own research projects.
Please RT.
Deadline: 11/09/2025
Details 👇:
shorturl.at/iiiOv
18.08.2025 05:35 — 👍 4 🔁 8 💬 0 📌 0

If you want to discuss #networks in #evo-devo, you should join this year's satellite symposium of the @dzg2025berlin.bsky.social organized by the developmental biology section (@marketa-kau.bsky.social, Benjamin Naumann, Alexander Klimovich). I will talk about gene regulatory networks.
08.08.2025 06:50 — 👍 9 🔁 5 💬 1 📌 0
116 FB 5 Research Assistant (m/f/d) field of Theoretical Ecology and Evolution or Computational Biology: Uni Osnabrück
I am excited to announce that the position of a senior postdoc (3 +3 years) in the field of theoretical biology is available in my group.
The position provides the opportunity to closely interact with experimentalists and develop own research projects.
Please RT.
Details 👇:
shorturl.at/iiiOv
08.08.2025 10:09 — 👍 95 🔁 119 💬 2 📌 4
OrthoFinder just dropped a major update
It’s faster, more accurate, and ready for thousands of genomes
Let’s break it down (1/10)
github.com/OrthoFinder/...
www.biorxiv.org/content/10.1...
16.07.2025 17:51 — 👍 126 🔁 73 💬 1 📌 1
🚨 Abstract deadline approaching!
🧬 Are you passionate about decoding life through genomics and cutting-edge science?
📅 Don’t delay—register today and submit your abstract!
🚨LINK to apply 👉 bit.ly/4n9AuqN
23.06.2025 15:32 — 👍 6 🔁 3 💬 0 📌 0
🧬 Reports mutations that give rise to coding potential & key properties of neORFs.
🛠️ Developed by Anna Grandchamp
🐳 Easy to run via Docker or Apptainer — no messy installs!
📄 Preprint: doi.org/10.1101/2025...
💻 GitHub: github.com/AnnaGrBio/DE...
🐋 DockerHub: hub.docker.com/r/edohmen/de...
12.06.2025 09:50 — 👍 0 🔁 0 💬 0 📌 0
🚨 New preprint alert! 🚨
🔬 Ever wondered how new genes emerge from scratch? Meet DESwoMAN, a fully automated pipeline to detect and analyze newly expressed ORFs (neORFs) from transcriptome data — giving us a window into the earliest stages of de novo gene emergence!
(Thread 🧵👇)
12.06.2025 09:50 — 👍 7 🔁 3 💬 1 📌 1

📖 SMBE journals on Bluesky
Interested in learning the latest advances in evolutionary biology, genomics, and molecular biology? Follow the SMBE sister journals MBE and GBE here on Bluesky and get updates on new articles coming out.
MBE: @molbioevol.bsky.social
GBE: @genomebiolevol.bsky.social
18.04.2025 07:32 — 👍 10 🔁 8 💬 0 📌 1
YUP. Made an error in the very first post. Correct link is: github.com/mol-evol/gcua
01.06.2025 19:31 — 👍 15 🔁 5 💬 1 📌 0
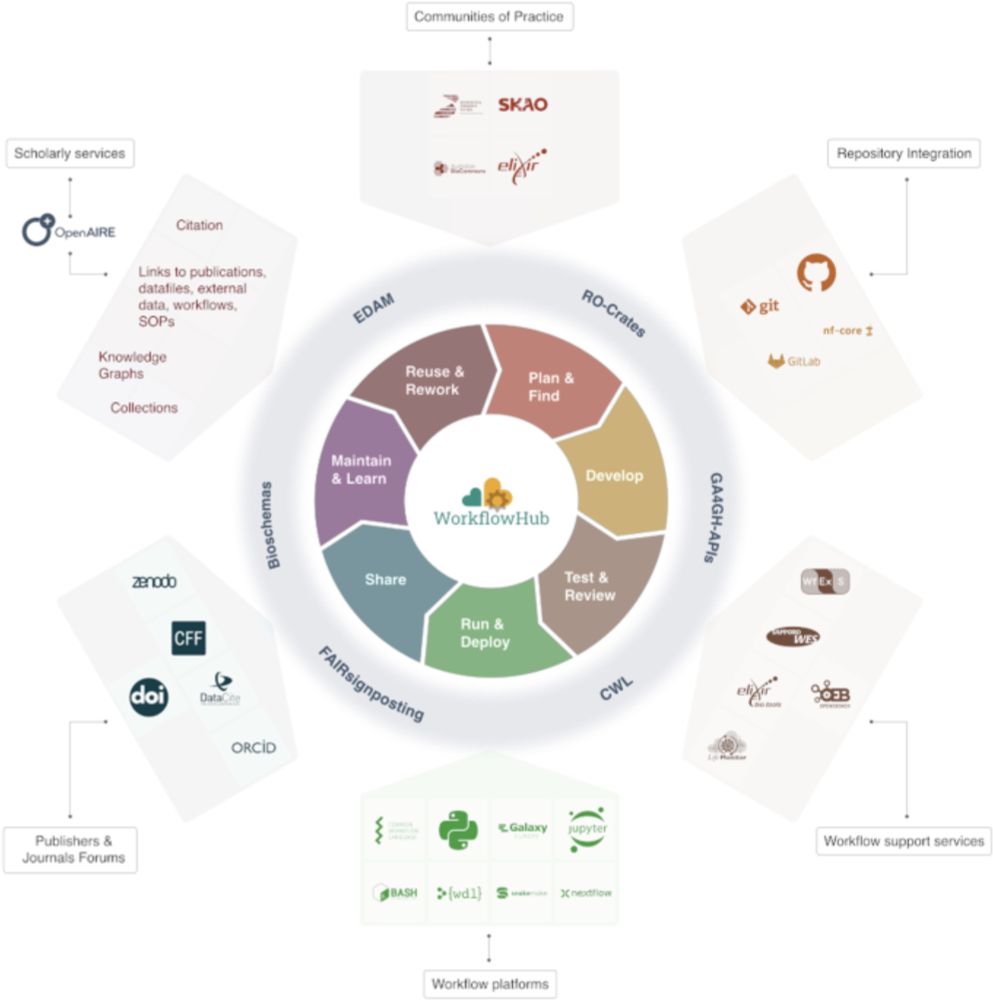
WorkflowHub: a registry for computational workflows - Scientific Data
Scientific Data - WorkflowHub: a registry for computational workflows
The WorkflowHub registry has a global reach, with hundreds of research organisations involved, and more than 800 workflows registered. www.nature.com/articles/s41... #bioinformatics #FAIR
24.05.2025 12:02 — 👍 8 🔁 7 💬 0 📌 0


Welcoming @lassemiddendorf.bsky.social as Mayor of iGEM’s Biomanufacturing Village.
This Village explores how we can grow the medicines, materials and commodities of the future using synthetic biology- powered by shared tools, scalable ideas and practical roadmaps.
villages.igem.org
16.05.2025 09:52 — 👍 5 🔁 3 💬 0 📌 0
Evolutionary Genomics Researcher
Adaptation, Transposon Elements, and Population Genetics.
University of Münster, Germany
- M.sc. student of Biosciences
- University Münster, Germany
- he/ him
Junior Professor for population genomics at University Hamburg https://www.biologie.uni-hamburg.de/forschung/populationsgenomik.html. Daphnia, Evolution and Data Science.
Neuroscientist studying tastes, smells, and the body-brain axis • New York-based researcher with faculty appointments at Nathan Kline Institute | NYU School of Medicine | City College of New York
nki.rfmh.org/people/renee-hartig-ph-d/
Associate Professor of Biological Sciences. We study genomes (WGD, etc.), and the ecology and evolution of fishes.
Evolutionary biologist, gene expression evolution, population genetics and fireflies
stochastic modelling in biology, evolutionary bioinformatics, theoretical population genetics, evolutionary (epi)genomics and transcriptomics
Structural Phylogenetics • methods, datasets, tools, events • concise updates for researchers and practitioners. Managed by @proteinmechanic.bsky.social and @cpuentelelievre.bsky.social
Postdoc in Claudia Bank’s group at the University of Bern. Promoters, emergence, fitness landscapes, dogs 🐶 and triathlon 🏊 🚲 🏃. He/him 🏳🌈.
timothyfuqua.com
Bioinformatician | Evolutionary Genomics | Microbiologist | Sex chromosome evolution
Senior Researcher at
@icrawater.bsky.social💧
Focus on #microbiology, #AMR, #OneHealth, #biofilms, #phages, #metagenomics, and #bioinformatics
Scientist Senckenberg Biodiversity and Climate Research Center, Frankfurt, Germany
evolutionary biology, adaptive genomics, behavior
Incoming DDLS Assistant Professor, Data Science and AI
division, Chalmers University | Formerly at JHU-LangmeadLab, UNIL-DessimozLab, WUR-deRidderGroup | Pangenomics for studying genome variation and evolution | Hiring PhDs/Postdocs CGRLab.github.io
Microbiome ecology, animal behaviour, evolutionary biology.
Postdoc at NIOO-KNAW
Previously: Postdoc at U. Oxford & PhD in EvoBio at U. Zurich.
Birder & bird ringing trainee. I dig Mini & cooking. He/him
Theoretical biologist
Postdoc @mainzuniversity.bsky.social
Evolution & development, gene regulation, health & disease, symbiosis
And a fondness for the humanities
https://scholar.google.com/citations?user=Sz-2qz4AAAAJ&hl=en
Full stack Dev at LoyJoy. 🌻
OrthoFinder
Evolution of cooperation
University of Oxford
https://scholar.google.com/citations?user=e3c1TdffPOcC
Postdoc Max Planck for Biological Intelligence and Max Delbrück Center, Berlin. Evolutionary biology, comparative and functional genomics. Birds, mammals. 🇲🇽🇩🇪🏳️🌈












