New paper by Amy Shipley and Lydia Woods
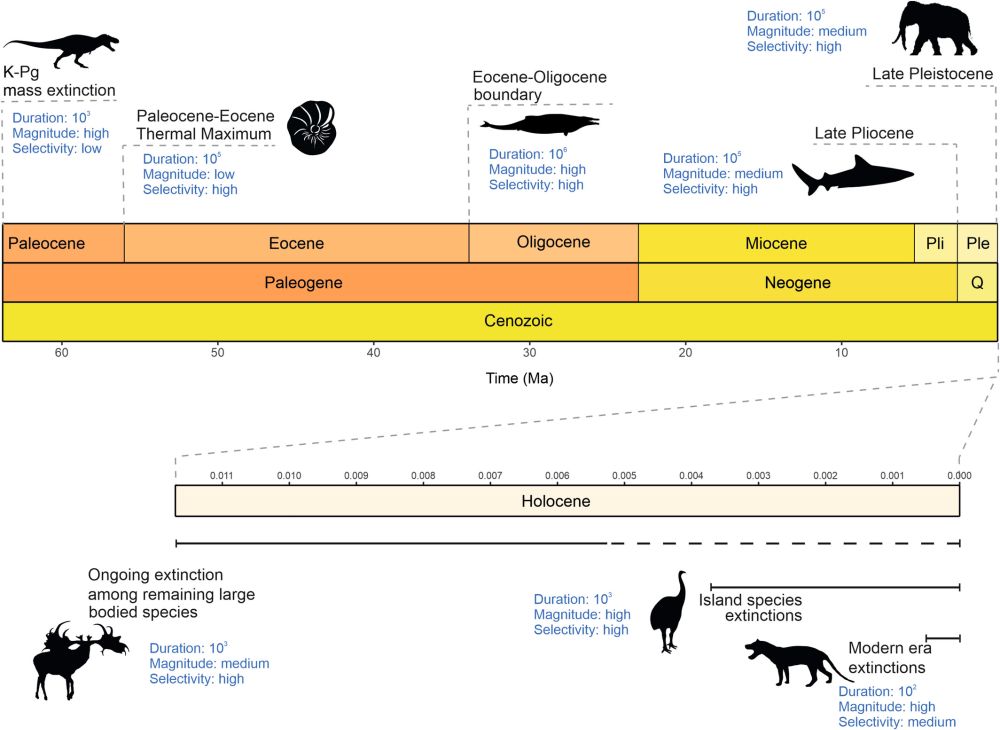
DeepBio@Leeds PGRs Amy Shipley and Lydia have published a review on the sixth mass extinction, as part of a working group on Cenozoic extinctions led out of the Anthropocene Biodiversity Centre at …
PGRs Amy Shipley (@sauropodlets.bsky.social) & Lydia Woods recently published a review in @globalchangebio.bsky.social showing that while today’s extinction rates aren't yet at "Big Five" mass extinction levels, they’re likely the highest seen in the last 66myrs.
amdunhill.co.uk/2025/11/17/n...
17.11.2025 13:46 — 👍 9 🔁 4 💬 0 📌 0

Out now in Biology Letters, my latest paper tackles an apparently simple question: how many characters are needed to reconstruct a phylogeny? TL;DR: in most cases between 100 and 500, more than a substantial portion of morphological datasets, but the story is more complex... doi.org/10.1098/rsbl...
15.10.2025 09:28 — 👍 88 🔁 48 💬 1 📌 1
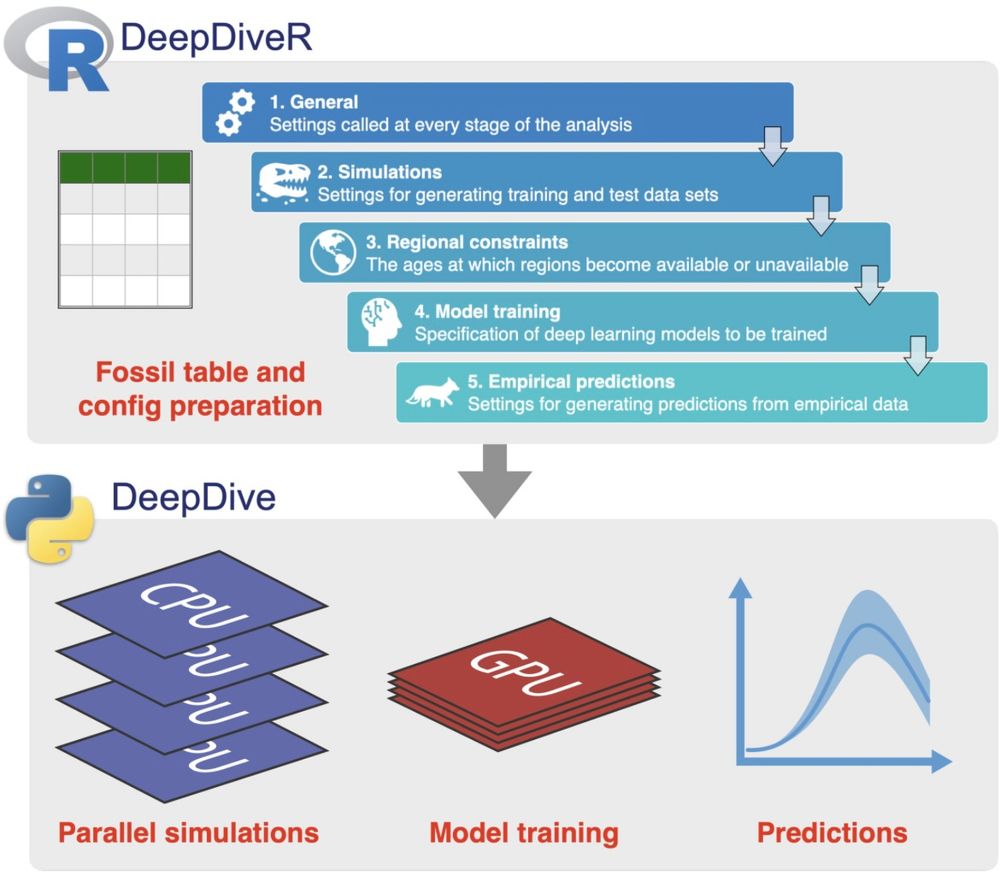
Really happy to see chapter 2 of my PhD published today in @methodsinecoevol.bsky.social. DeepDiveR is an R package to assist with deep learning inference of biodiversity change through time 🤿🦊 Thanks to @bethanyjallen.bsky.social & Daniele Silvestro for the collaboration
doi.org/10.1111/2041...
08.08.2025 10:44 — 👍 29 🔁 13 💬 1 📌 0
A covarion model for phylogenetic estimation using discrete morphological datasets https://www.biorxiv.org/content/10.1101/2025.06.20.660793v1
27.06.2025 04:32 — 👍 6 🔁 2 💬 1 📌 0

A woman delivering a talk
@lauramulvey.bsky.social absolutely killing it talking about the difference between model adequacy and model selection and describing lightweight methods to assess adequacy. #evol2025
Paper here academic.oup.com/sysbio/artic...
22.06.2025 14:13 — 👍 25 🔁 3 💬 1 📌 0

European Australian lifts leg onto window sill and holds aloft a Workshop invitation.
Sometimes in a fossil policy workshop you have to take a strong stand.
@thepalass.bsky.social
16.05.2025 09:58 — 👍 4 🔁 1 💬 0 📌 0
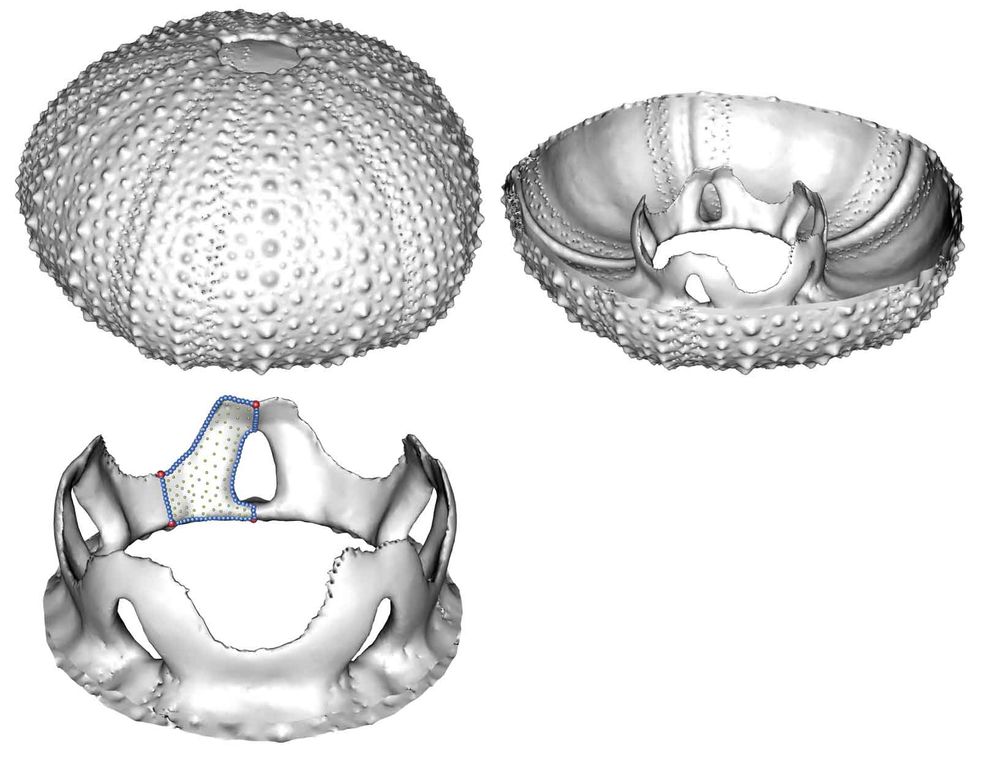
Interested in Developmental biology? Imaging? Marine invertebrate body plans? Then this three year postdoc in my group @sotonbiosciences.bsky.social is for you! Join our team to decypher how signalling molecules shape skeletal phenotype in juvenile sea urchins. jobs.soton.ac.uk/Vacancy.aspx...
12.05.2025 11:00 — 👍 44 🔁 30 💬 1 📌 3
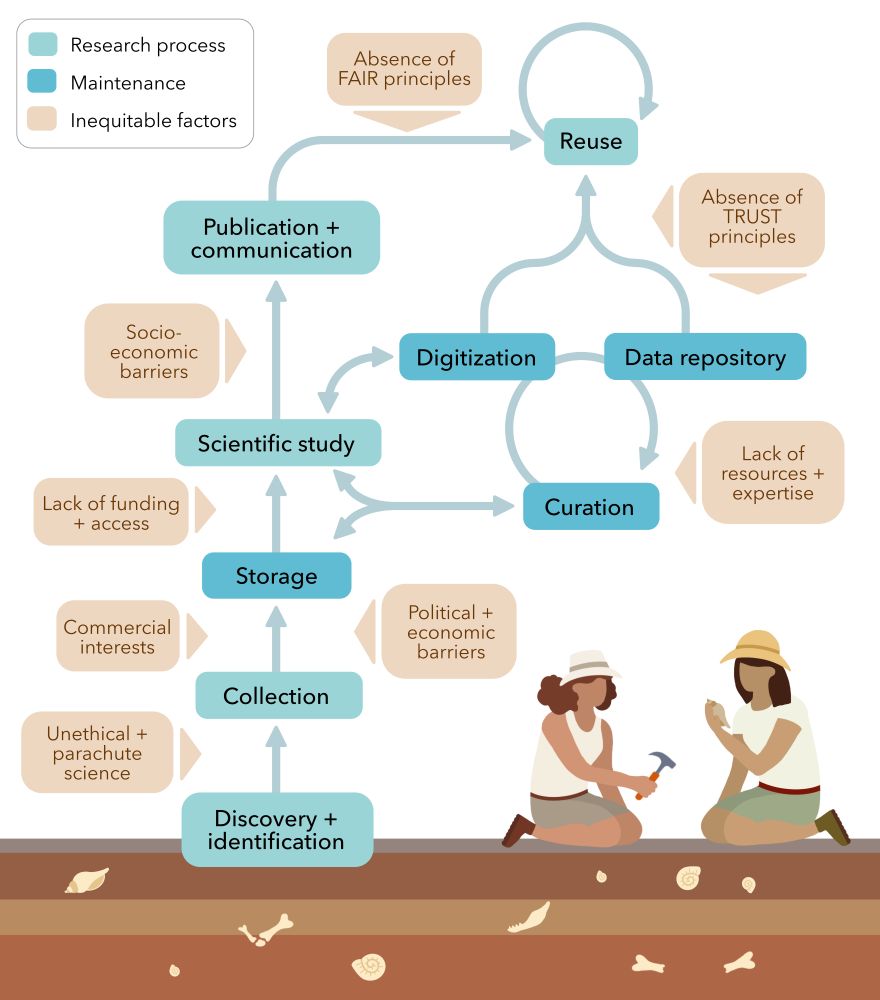
Simplified flowchart illustrating generalised steps in palaeobiological research processes and the various factors that introduce inequity with regard to data collection, storage, study, analysis, publication, and reuse.
Delighted to share our paper on data equity in #palaeobiology as part of Paleobiology's 50th anniversary issue 🥳
We look at how palaeo data is collected, stored, curated & shared, and how equity in these processes is crucial for our field's future (1/n) 🧪⚒️
doi.org/10.1017/pab....
23.04.2025 13:01 — 👍 101 🔁 42 💬 3 📌 2
📢 Tired of choosing between fossils to incorporate into your phylogenetic analysis? Us too.
💥 Check out our new preprint on incorporating stratigraphic ranges into the FBD model, implemented in BEAST2. We explore how using ranges affects the inference of dog 🐶 and penguin 🐧 trees 🌳
24.04.2025 08:57 — 👍 38 🔁 21 💬 2 📌 1
From fossils to phylogenies: exploring the integration of paleontological data into Bayesian phylogenetic inference | Paleobiology | Cambridge Core
From fossils to phylogenies: exploring the integration of paleontological data into Bayesian phylogenetic inference - Volume 51 Issue 1
🎂 Happy 10th birthday to the fossilised birth-death model, and happy 50th birthday to Paleobiology 🎂
I had a lot of fun working on this review, led by @lauramulvey.bsky.social, in which we celebrate how the FBD has been applied and give pointers to those interested in giving it a go! 🌳
23.04.2025 08:23 — 👍 55 🔁 21 💬 0 📌 1
Combining fossil taxa with and without morphological data improves dated phylogenetic analyses https://www.biorxiv.org/content/10.1101/2025.04.07.646048v1
09.04.2025 15:32 — 👍 13 🔁 10 💬 0 📌 3
YouTube video by The Palaeontological Association
Palaeontologists Explain: How do fossils form? Palaeontologists explain: How do fossils form?
Very proud to be featured alongside other excellent early career scientists in a new video by @thepalass.bsky.social!
We talk about how fossils form and how rare they are!
youtu.be/LwF7qnYFde8?...
25.03.2025 19:06 — 👍 38 🔁 20 💬 4 📌 0
The clock is ticking 🕔! Pre-registration will close today March 14th (17:00h GMT+0), do not miss your chance to join us at the SystAssn-SRUK/CERU Bayesian Phylogenetics Workshop! 💻🧬
14.03.2025 14:24 — 👍 2 🔁 1 💬 0 📌 0
Like morphometrics, micro-CT scanning, and morphological evolution? Then please consider applying for our 18 month postdoc position at the university of Southampton! Details here: www.jobs.ac.uk/job/DMF402/r...
12.03.2025 17:17 — 👍 48 🔁 44 💬 2 📌 0
And finally have it in preprint: www.biorxiv.org/content/10.1...
In it, we (@basantakhakurel.bsky.social, @brenenwynd.bsky.social, @peterjwagner3.bsky.social, and Christian Kammerer) took a look at estimating a phylogenetic tree of dicynodonts using both discrete and continuous characters.
07.03.2025 15:45 — 👍 44 🔁 16 💬 2 📌 1

As my very first post here on Bluesky I am happy to share our article in Royal Society about tooth wear and diet in the large Devonian chondrichthyan Ctenacanthus concinnus from Morocco
royalsocietypublishing.org/doi/10.1098/...
29.01.2025 08:09 — 👍 14 🔁 4 💬 0 📌 0
Come be my colleague! Calling zoologists of all stripes! This is a pretty open search! If you research is cool and forward looking and if you care about good science questions and collections please apply!
13.01.2025 19:54 — 👍 15 🔁 15 💬 1 📌 1

A reminder that we have two open PhD projects on Bayesian phylogenetics in the lab:
One with the new TREE Doctoral Landscape Awards: www.trees-dla.ac.uk/projects/int... (deadline 20th Jan)
And another with CSC: www.findaphd.com/phds/project... (deadline 29th Jan)
06.01.2025 19:42 — 👍 26 🔁 29 💬 0 📌 2
🚨New #phylo paper alert!🚨 🌲+🔥= speciation? This one has been a bit of a journey, but thanks to lead author Dan Turck's expertise on #conifers it has turned out as quite the treasure-trove of conifer evolution! (reading-access link on my website)
05.01.2025 19:55 — 👍 28 🔁 14 💬 0 📌 0
Assessing the impact of character evolution models on phylogenetic and macroevolutionary inferences from fossil data https://www.biorxiv.org/content/10.1101/2024.12.23.630137v1
24.12.2024 05:35 — 👍 14 🔁 6 💬 0 📌 0

A normal distribution drawn on graph paper decorated like a Christmas tree. Along the x-axis it says happy Xmas
Happy Christmas!
24.12.2024 10:02 — 👍 54 🔁 6 💬 2 📌 0
RevBayes: Assessing Phylogenetic Reliability Using RevBayes and $P^{3}$
Check out the tutorial for posterior predictive simulations on the revbayes website
revbayes.github.io/tutorials/pp...
20.12.2024 09:36 — 👍 8 🔁 3 💬 0 📌 0
Huge thank you to all my co-authors for their help and guidance throughout this project @rachelwarnock.bsky.social, @wrightam.bsky.social, @hoehna.bsky.social, @jembrown.bsky.social, and Mike May 6/n
20.12.2024 09:36 — 👍 7 🔁 0 💬 1 📌 0
Further, we show how model selection approaches relying on calculating the marginal likelihoods, cannot be used to distinguish between models with different partitioning schemes. 5/n
20.12.2024 09:36 — 👍 5 🔁 0 💬 1 📌 0
We found that for many (but not all) empirical data sets, available models were in fact adequately describing the evolutionary process, and were therefore suitable for inference. 4/n
20.12.2024 09:36 — 👍 5 🔁 0 💬 1 📌 0
We adopted a model adequacy approach to assess the absolute fit of these models, and developed a workflow for posterior predictive simulations to evaluate how well available models of morphological evolution fit empirical data. 3/n
20.12.2024 09:36 — 👍 5 🔁 0 💬 1 📌 0
Macroevolution, macroecology, phylogenetics, palaeontology, beer taxonomy and unplanned stuff. He/him. Mainly on @TGuillerme@mastodon.social
Palaeontologist specialising in the behaviour of dinosaurs and biology of pterosaurs. Reader in Zoology @QMUL. Author of several books, podcaster, general sci-commer. Thoughts my own. He / him. davehone.co.uk
Postdoctoral researcher at LMU | macroevolution of crocodyliforms 🐊
BA in Environmental Biology, MSc in Geobiology and Paleobiology, expert in googling
A free pre-print and post-print service for ecology, evolution, and conservation. Run by @sortee.bsky.social.
Server: https://ecoevorxiv.org
Instructions & FAQ: https://sortee.github.io/ecoevorxiv/
MSc @LMU Munich. Method development in phylogenetics, currently focusing on comparative methods
Macroevolutionary Palaeobiology Postdoc | Fritz Lab (BinA), German Centre for Integrative Biodiversity Research (iDiv) | Fossil Mammals ❤
An international, peer-reviewed, life science journal, devoted to whole-organism biology. Edited and co-published by the Leibniz Institute for the Analysis of Biodiversity Change and Pensoft.
Website: evolsyst.pensoft.net
Postdoc at SBBS, QMUL, London. Evolutionary Biologist. Interested in the evolutionary mechanisms of host microbes interactions. @QM_SBBS @QMUL @MPI_Evolbio
Palaeontologist, invertebrate botherer. PhD student at the University of Bristol. Interested in the evolution of Ecdysozoa. Mainly working on worm fossils from the Cambrian Sirius Passet Lagerstätte.
Paleobiology, trilobites and disco | Postdoc at Stanford University
Vertebrate paleontologist and evolutionary biologist, currently postdoc at LMU Munich. In a love-hate relationship with phylogenies :D
Bioinformatics, Statistical Phylogenetics, molecular evolution,
Eco-Evo researcher, PhD from Queen Mary University of London,
Plant enthusiast and nature lover | he/him
Scientist, Evolutionary Biologist & Entomologist / Ants / Genomes to Microbiomes / Professor & Curator at @Cornell / @CornellCALS / She/Her
postdoc @floridamuseum.bsky.social phenomics of insect evolution. toward AI/CV-based museomics. www.luerig.net
Twin, entomologist, evolutionary biologist 🇨🇦🏳️🌈🇺🇸
🇧🇷 Computational paleobiology, macroevolution, Bayesian phylogenetics, and canids 🐶
PhD candidate @ Iowa State University
brpetrucci.github.io
evolutionary biologist (phylogenetic network methods, plant systematics) | postdoc @ University of Washington | PhD from University of Wisconsin-Madison | 🏃🏻♀️✨🌈🌻🧬 she/her
Botanist. Evolutionary biologist. Associate Professor and Herbarium Director, LSU. Passionate about plants, inclusive mentoring, and puzzles. I don't know what I'm doing here.
https://www.lagolab.net













