Europe is stepping up, helping Ukraine for the right moral reasons and helping their mutual economies.
27.10.2025 13:55 — 👍 11 🔁 1 💬 0 📌 0

BMP signaling-dependent dorsal-to-ventral gradient of PAX3/7 transcriptional activity revealed by the P34::tk::LacZ reporter. Left: expression patterns of neural progenitor (NP) and interneuron (IN) TF markers in the developing spinal cord. Right: Immunostaining for β-galactosidase (β-gal; red/grey), GFP (blue/grey) and either OLIG3 (green/grey) or pSMAD1/5/9 (green/grey) on transverse sections of E9.0 (bottom) and E9.5 (top) P34::tk::LacZ; Pax3+/GFP spinal cords at brachial level. Black and white panels show magnified views of the boxed region.
How do pleiotropic TFs generate organized diversity in developing tissues? @spinalorga.bsky.social shows that PAX3 & PAX7 organize #SpinalCord by acting as both repressors & pioneer activators, regulated by #morphogens to ensure precise neural subtype specification @plosbiology.org 🧪 plos.io/4qumsla
27.10.2025 17:25 — 👍 20 🔁 10 💬 0 📌 0

Cracking the code of the non-coding genome via allele-specific genomics?
Can we link non-coding elements—like lncRNAs and enhancers—to their protein-coding target genes, and in doing so, connect overlapping non-coding disease variants to their protein-coding counterparts?
24.10.2025 02:43 — 👍 28 🔁 12 💬 2 📌 1
On Sunday I will cycle 50 km to raise money to support research to prevent blindness. If you can, please sponsor me! fightingblindness.akaraisin.com/ui/MOVEFORSI...
20.06.2025 17:37 — 👍 10 🔁 4 💬 0 📌 0

World first: ultra-powerful CRISPR treatment trialled in a person
Immune-cell function improved in a teenager whose DNA was altered using prime editing.
The CRISPR family’s most versatile member has made its medical debut: a cutting-edge gene-editing technique known as prime editing has been used to treat a person for the first time
https://go.nature.com/3YNta9V
19.05.2025 13:58 — 👍 71 🔁 19 💬 0 📌 1

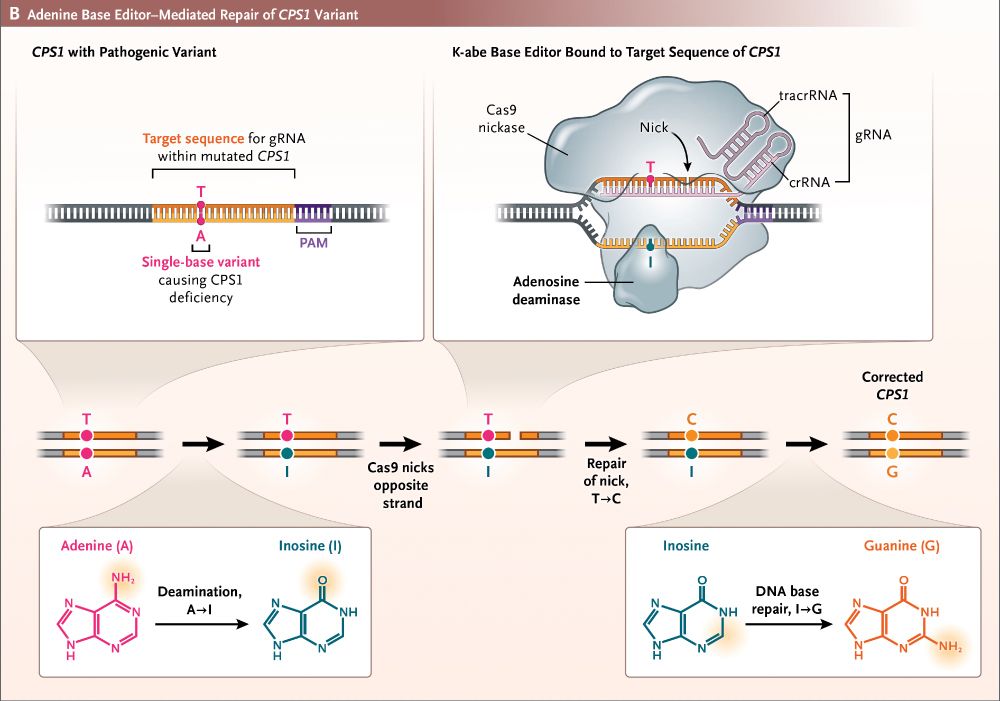
Today "a milestone in the evolution of personalized therapies for rare & ultra-rare inborn errors of metabolism"
—the 1st human to undergo custom genome editing
—from decades of NIH funded research
www.nejm.org/doi/full/10....
@nejm.org
www.nejm.org/doi/full/10....
www.nytimes.com/2025/05/15/h...
15.05.2025 18:05 — 👍 596 🔁 169 💬 10 📌 27
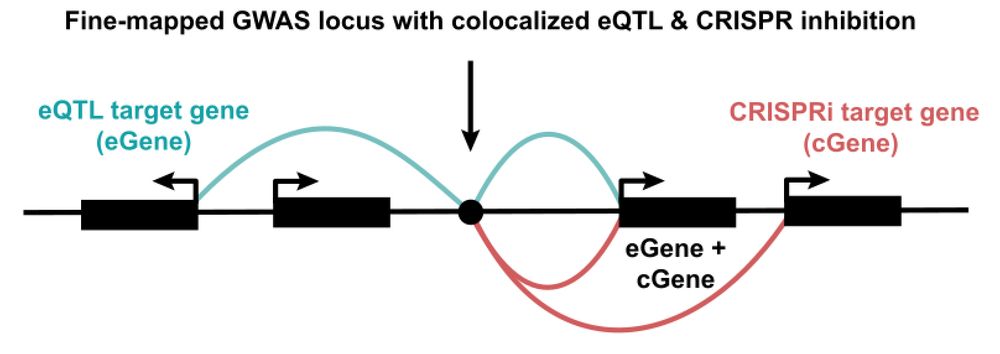
Our new contribution to the quest to find causal GWAS genes! Sam Ghatan from my lab at @nygenome.org led a systematic comparison of eQTLs and CRISPRi+scRNA-seq screens. TL;DR: they provide highly complementary insights, with ortogonal pros and cons. 🧵👇
www.biorxiv.org/content/10.1...
06.05.2025 17:00 — 👍 98 🔁 42 💬 1 📌 2

Vilcek Prizes for Creative Promise - Vilcek Foundation
The Creative Promise Prizes are awarded to young immigrant professionals who have demonstrated outstanding achievement early in their careers.
If you are immigrant into the US and work in the biomedical sciences (like me) and are still young (not like me), please apply for the Vilcek prizes for Creative Promise founded by an inspiring immigrant couple, Marica and Jan Vilcek.
vilcek.org/prizes/vilce...
18.03.2025 13:45 — 👍 16 🔁 10 💬 1 📌 0
🚨 New paper alert! 🚨
We’ve just launched openretina, an open-source framework for collaborative retina modeling across datasets and species.
A 🧵👇 (1/9)
14.03.2025 09:41 — 👍 38 🔁 20 💬 1 📌 1

Saying goodbye is hard! So is interpreting non-coding variants in visual disorders! Luckily, that is the gift that Dr. Leah VandenBosch left us with after a very successful postdoc in the Cherry Lab. Check your own favorite retinal regulatory variants here: genome.ucsc.edu/s/CherryLab/...
26.02.2025 02:49 — 👍 9 🔁 3 💬 1 📌 1
#Neuroskyence students! We're brainstorming ways to support your development over this summer break, esp. given the changes in summer intern programs across our fields. What other opportunities would you like to see?
Some ideas: undergrad-focused webinars, self-paced courses, challenges, etc.
25.02.2025 17:01 — 👍 30 🔁 10 💬 3 📌 2
I was touched by the great turnout in Seattle to support #Ukraine. Thank you to the organizers and @adamsmith.house.gov for your speech. We must continue arming Ukraine and defend democracy against Russian invasion. #StandWithUkraine 🇺🇦"
24.02.2025 02:33 — 👍 5 🔁 0 💬 0 📌 0
Since 2022, people around the world have stood in solidarity, joining rallies to support Ukraine and refusing to let russia silence the truth. They chose Ukraine’s side, united in the fight for freedom. Join in, speak up, #StandWithUkraine:
uaworldmap.ukrainianworldcongress.org/en?fbclid=PA...
23.02.2025 16:08 — 👍 176 🔁 38 💬 3 📌 1
Your city might have separate events, like the one I'll be attending in Seattle, organized by the Ukrainian Association of Washington State
21.02.2025 20:25 — 👍 2 🔁 0 💬 0 📌 0

Global Events Where You Can Stand With Ukraine as Russia’s Full-Scale War Hits the Three-Year Mark
Global events mark three years since Russia's invasion of Ukraine, showcasing solidarity and raising awareness for ongoing support and resilience.
As Russia's full-scale war hits the three-year mark, it's more important than ever to #StandWithUkraine. I encourage you to raise your voices for peace and justice by attending one of the events organized worldwide.
#RussiaIsATerroristState
21.02.2025 20:24 — 👍 1 🔁 0 💬 1 📌 0
Negotiating with the murderous dictator Putin won't lead to anything. He needs to be forced to retreat from Ukraine.
18.02.2025 18:52 — 👍 1 🔁 0 💬 0 📌 0
I hope this hastens the fall of Putin's regime
28.01.2025 21:04 — 👍 1 🔁 0 💬 0 📌 0
Since Russia's invasion of Ukraine, almost 20,000 Ukrainian children have been kidnapped by Russia. DNA testing might be the only way to one day reconnect them with their families.
22.01.2025 04:28 — 👍 6 🔁 0 💬 0 📌 0
I'm sure the Russian government will deny everything similar to MH17 and countless wat crimes they commit in Ukraine.
26.12.2024 16:10 — 👍 1 🔁 0 💬 0 📌 0

As a result of this morning's shelling, 630 residential buildings, 16 medical facilities, 17 schools, and 13 kindergartens have been left without heat.
We can’t fully protect🇺🇦schools from russian missiles, but we can #BringLightBack to them:
u24.gov.ua/bring-light-...
20.12.2024 12:32 — 👍 178 🔁 78 💬 3 📌 3
A new interesting study investigating mitochondrial DNA (mtDNA) cis-effects on mtDNA encoded genes, mtDNA trans-effects on nucDNA encoded genes, and nucDNA trans-effects on mtDNA encoded genes.
#genomics #generegulation
11.12.2024 06:17 — 👍 1 🔁 0 💬 0 📌 0
An exciting preprint from Choudhary Lab addressing how cohesin and CTCF control gene expression and their role in precise enhancers targeting.
Cohesin appears to facilitate enhancer-mediated gene regulation via enhancers that depend on p300/CBP.
11.12.2024 03:35 — 👍 1 🔁 0 💬 0 📌 0
JAMA Ophthalmology is a member of the JAMA Network, a consortium of peer-reviewed, general medical and specialty publications.
🌐 JAMAOphthalmology.com
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
PhD Student @ UW Genome Sciences
🧬 PhD Candidate | Molecular and Cellular Biology | University of Washington | Long Read Sequencing and Structural Variants | Diversity and Equity in STEM | he/him 🏳️⚧️
PhD candidate at UW Seattle Molecular and Cellular Biology program 🧬 Studying retinal development and disease 👁️
Developing advanced imaging techniques to study how the neurons in the retina talk to each other. PhD student at @sln.icfo.eu
We’re advancing pediatric health through groundbreaking research. As a top 5 pediatric research center and top 10 pediatric hospital in the US, we’re providing hope, care and cures to help every child live the healthiest and most fulfilling life possible.
Postdoc @ Kaessmann and Sasse labs @zmbh.uni-heidelberg.de
Previously PhD @ Sebé-Perdós lab @crg.eu
Interested in regulatory genomics, evolution, machine learning, and especially the combination of all of the above.
https://anamaria.elek.hr/
🦋 #Vision #science| #butterfly wrangler | Distinguished Professor | #Guggenheim | American Academy of Arts and Sciences | NAS | Words: NPR, PBS, ConversationUS | Macondista | Interested in #history | Latina in #STEM
Lab website: www.visiongene.org
MD/PhD Student in Doug Fowler's Lab, UW Genome Sciences
Professor in the Department of Cell and Developmental Biology in the School of Biological Sciences at UC San Diego.
Director of the PhD program in Biological Sciences at UCSD and the Salk.
Asking the hard questions and bringing unique perspectives from across the globe. This is CNN.
Biology prof @ #PUI, former dept chair, fish guy, developmental biologist, and vision researcher. Here for the science, football and #FPL. #COYB
https://www.bodegacats.nyc
Group leader at MPI Molecular Biomedicine. Growing organoids and poking embryos to understand how functional organs are built.
https://www.mpi-muenster.mpg.de/researchgroups/rocha
Graduate Student in the Wert Laboratory - UT Southwestern Department of Ophthalmology. All views/opinions are my own
Phage’s mom, scientist, snow enthusiast
www.dramandagunn.com
Reporting from the frontiers of health & medicine.
The Institute for Stem Cell and Regenerative Medicine at the University of Washington is changing the way human beings confront chronic disease.