We are thrilled that our study on the evolution of gene regulation in mammalian cerebellum development – led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social – is now out in @ScienceMagazine! www.science.org/doi/10.1126/...
29.01.2026 19:23 — 👍 92 🔁 35 💬 3 📌 7

Exciting news to start the new year! We’re thrilled to see this work finally out in @natecoevo.nature.com, the result of a major collaboration between my lab and the labs of Veronica Hinman and Nacho Maeso, began many years ago with the dear José Luis Gómez-Skarmeta.
www.nature.com/articles/s41...
07.01.2026 10:17 — 👍 40 🔁 16 💬 1 📌 3
Science: only when you write up the manuscript, you realize what you should have done.
16.09.2025 15:56 — 👍 167 🔁 32 💬 7 📌 5

We are happy to share our latest work in @nature.com . We study the genomic and cellular basis of facultative symbiosis in Oculina patagonica - a Mediterranean coral remarkable for its ability to survive long periods without algal symbionts. Led by Shani Levy and @xgrau.bsky.social
rdcu.be/eLbaZ
15.10.2025 19:57 — 👍 133 🔁 48 💬 5 📌 2

Picture of an Oculina patagonica colony. Some of the individual polyps in the colony have a brown-yellow colour, indicating that they harbour symbiotic algae. Others are completely white and lack algae.

Close-up picture of symbiotic Oculina polyps.
Hot off the press! Our latest work on the evolution of facultative symbiosis in stony corals, focusing on a remarkable Mediterranean species: Oculina patagonica.
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
15.10.2025 20:29 — 👍 46 🔁 11 💬 2 📌 0
Excited to share the preprint from my main postdoc project! It’s been a long journey—huge thanks to everyone who made it possible, especially @leticiarm1618.bsky.social for being the best collaborator one could ask for, and the amazing @kaessmannlab.bsky.social lab for the invaluable support!
15.10.2025 13:39 — 👍 13 🔁 6 💬 1 📌 0
BCA is a project to look out for — charting the diversity of cell type transcriptomes across the tree of life. Not only will it empower evolutionary studies, but also drive advances in biotechnology, biomedicine, and ecology. Kudos to the relentless, meticulous, and persistent team doing this work!
25.09.2025 09:47 — 👍 6 🔁 1 💬 0 📌 0

🚀 Check out our new review article “From Tiny Exons to Big Insights: The Expanding Field of #Microexons” now out in Annual Review of Genomics and Human Genetics!
doi.org/10.1146/annu...
Special thanks to @mirimiam.bsky.social, @crg.eu and @upf.edu!
31.08.2025 13:42 — 👍 10 🔁 3 💬 0 📌 1
A shout-out to people who made this possible: first and foremost @martaig.bsky.social and @arnausebe.bsky.social, but also @zolotarg.bsky.social @xgrau.bsky.social as well as all the ASP lab members, and of course @lukasmahieu.bsky.social @steinaerts.bsky.social and all the members of LCB in Leuven
06.07.2025 18:14 — 👍 3 🔁 0 💬 0 📌 0
We anticipate that applying the same approaches to other species of cnidarians and early-branching animals will enable comparative cell type analyses that will reconstruct evolutionary relationships of the major animal cell types and regulatory processes by which they first evolved.
06.07.2025 18:14 — 👍 1 🔁 0 💬 2 📌 0
To wrap up, here we pave the way for moving beyond conventional transcriptome-based cell type characterization in non-model species, by analyzing regulatory traits that define cell type identities in Nematostella, such as CREs sequence motif composition, active TFs, and GRN architecture.
06.07.2025 18:14 — 👍 0 🔁 0 💬 1 📌 0
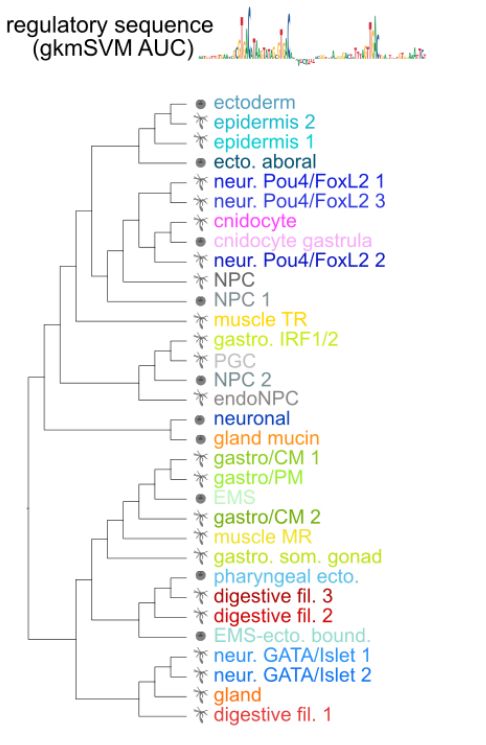
Cell type relationships based on regulatory sequence similarity
We therefore show that effector gene usage groups functionally similar cell types, but regulatory features also reflect their ontogenetic relationships. E.g. GATA/Islet neurons show regulatory seq. similarities with
EMS and pharyngeal derivatives, and Pou4/FoxL2 neurons with ectodermal derivatives.
06.07.2025 18:14 — 👍 0 🔁 0 💬 1 📌 0
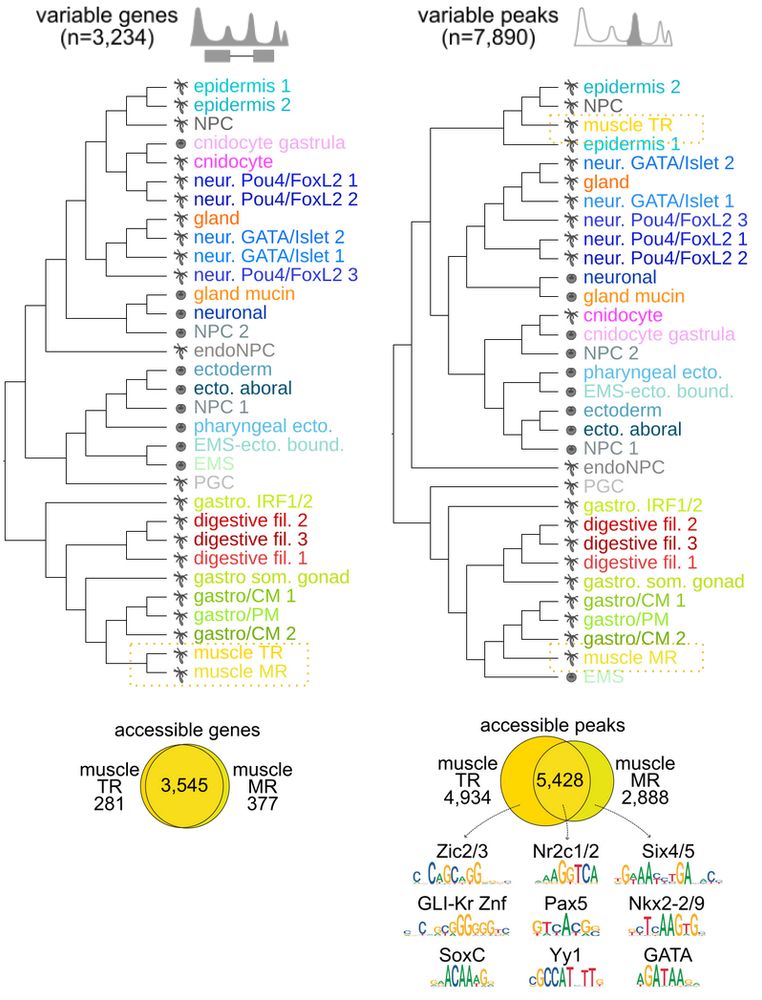
Finally, we explored cell type clustering using different features. We highlight transcriptionally similar retractor muscles, which share many access. genes, but have distinct sets of CREs bound by distinct TFs, and each clusters with the derivatives of their precursors (ecto. for TR and EMS for MR)
06.07.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0
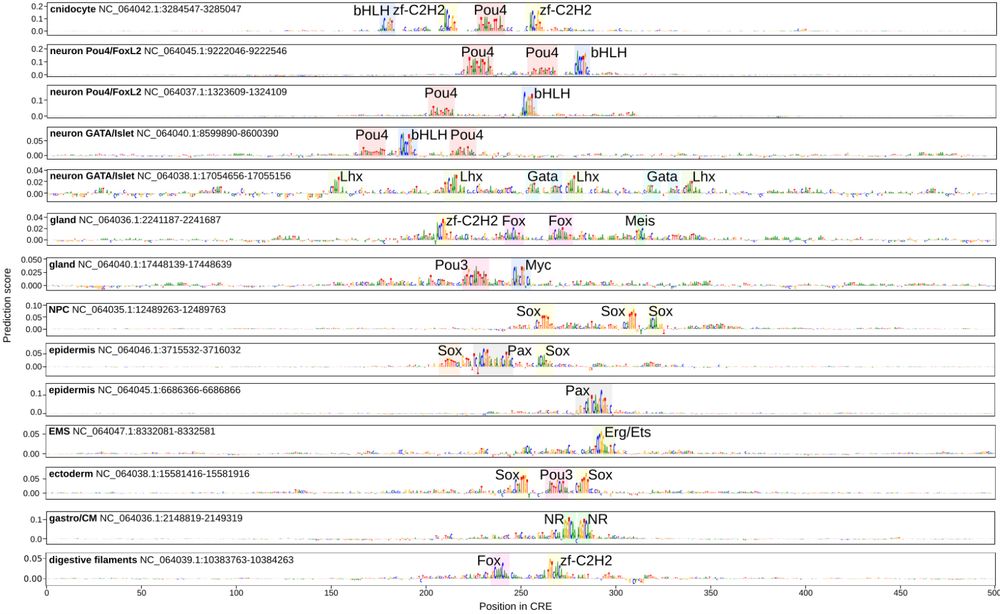
Motifs identified with deep learning in CREs of different cell types.
With invaluable help of @lukasmahieu.bsky.social and @steinaerts.bsky.social lab we trained deep learning sequence models to prioritize motifs predictive of cell type specific accessibility, and to uncover mostly flexible motif syntax in Nematostella, in line with billboard-like model of TF binding.
06.07.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0
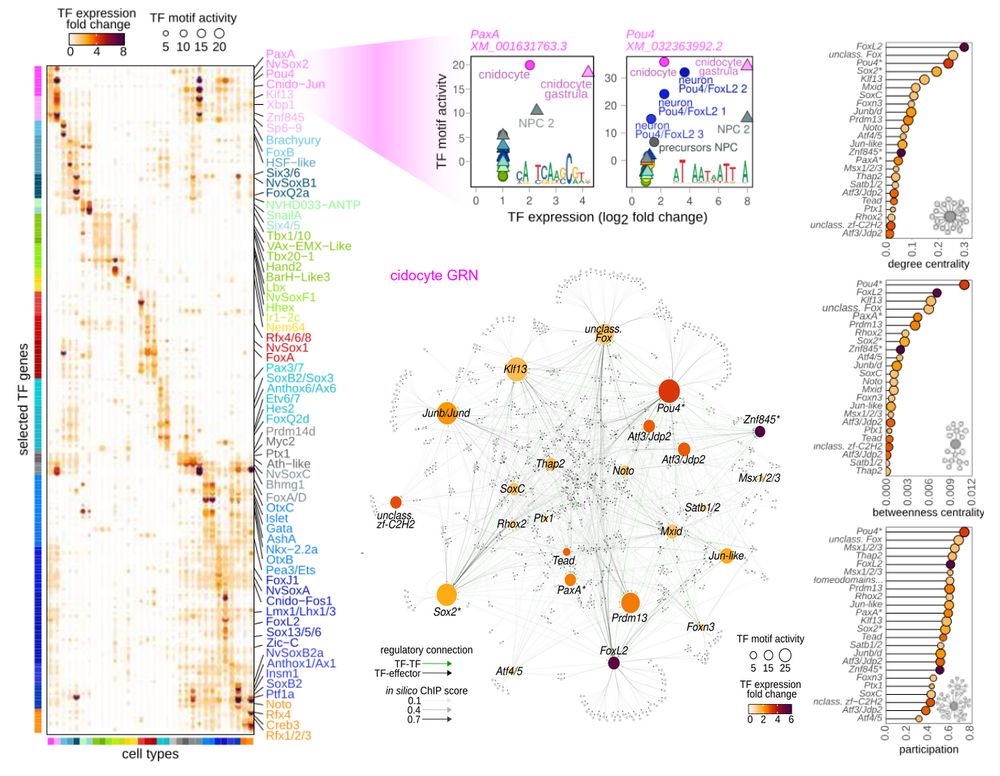
Cnidocyte gene regulatory characterization.
With that in hand, we characterized each cell type by usage of TF motifs, and then linked active TFs to their target genes in cell type specific gene regulatory networks (GRNs). We showcase cnidocyte GRN as an example and highlight important TFs with central roles in the network (FoxL2, Pou4, Sox2).
06.07.2025 18:14 — 👍 0 🔁 0 💬 1 📌 0
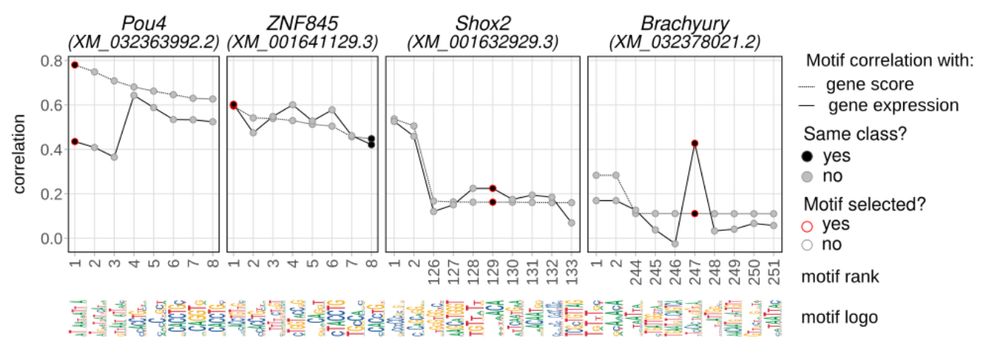
Correlation-based motif assignment approach.
The link between ATAC and RNA - from gene regulatory perspective - are TF binding motifs, which are not known for most Nematostella TFs. We devised a correlation-based approach to assign one motif to each TF, selected as best correlated among all motifs inferred by sequence similarity and orthology.
06.07.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0
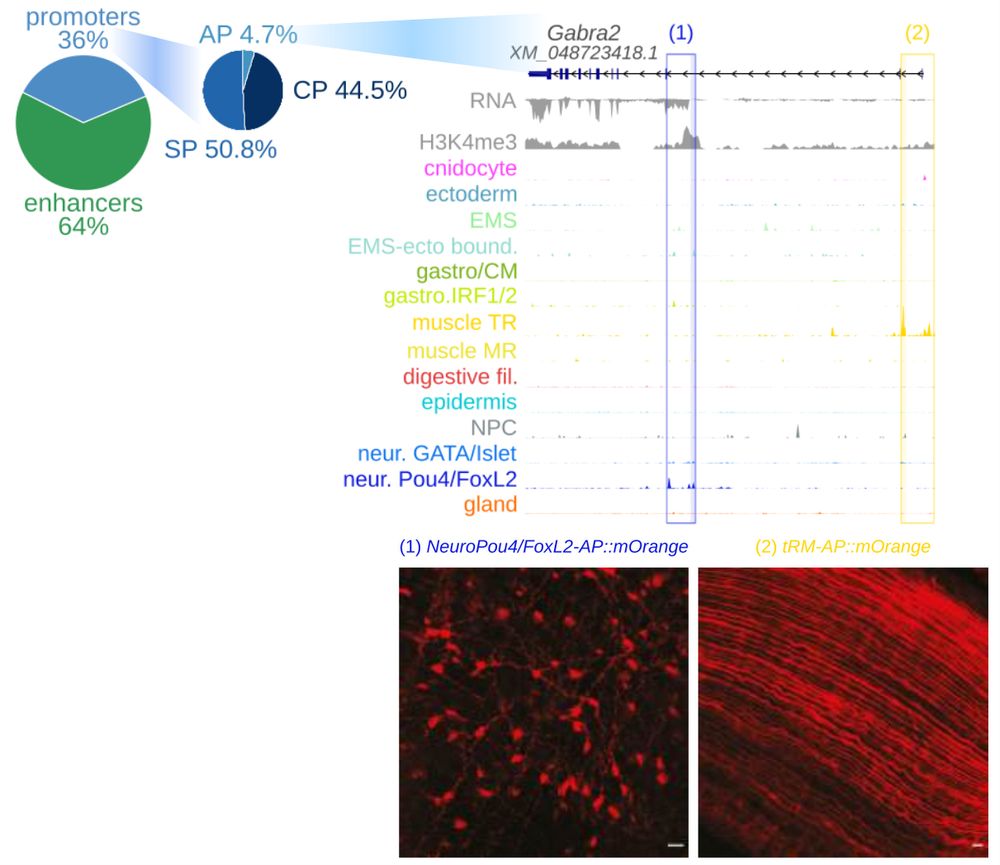
Alternative promoter of Gabra2 in neurons and muscle.
We used the atlas to characterize and quantify candidate CREs, including cell type-specific enhancers, cell type-specific promoters (SP), constitutive promoters (CP) and a smaller number of candidate alternative promoters (AP). We validated muscle and neuron AP of Gabra2 using transgenic reporters.
06.07.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0
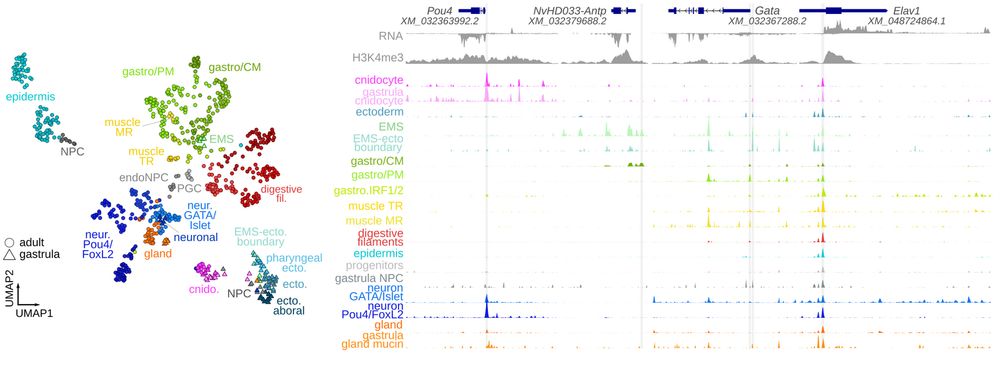
scATAC atlas of Nematostella vectensis.
To start, @martaig.bsky.social produced the first scATAC atlas for a non-model species, profiling 60k cells from adult and gastrula Nematostella vectensis - see it annotated in the app: sebelab.crg.eu/nematostella-cis-regulatory-atlas/ and the genome browser: sebelab.crg.eu/nematostella-cis-reg-jb2
06.07.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0
In this project we wanted to extend cell type characterization in early-branching animals from transcriptome-based (scRNA) to regulatory-based definition, by experimentally profiling chromatin accessibility (scATAC) and computationally inferring TF binding to cis-regulatory elements (CREs).
06.07.2025 18:14 — 👍 1 🔁 0 💬 1 📌 0
Decoding cnidarian cell type gene regulation
Animal cell types are defined by differential access to genomic information, a process orchestrated by the combinatorial activity of transcription factors that bind to cis -regulatory elements (CREs) to control gene expression. However, the regulatory logic and specific gene networks that define cell identities remain poorly resolved across the animal tree of life. As early-branching metazoans, cnidarians can offer insights into the early evolution of cell type-specific genome regulation. Here, we profiled chromatin accessibility in 60,000 cells from whole adults and gastrula-stage embryos of the sea anemone Nematostella vectensis. We identified 112,728 CREs and quantified their activity across cell types, revealing pervasive combinatorial enhancer usage and distinct promoter architectures. To decode the underlying regulatory grammar, we trained sequence-based models predicting CRE accessibility and used these models to infer ontogenetic relationships among cell types. By integrating sequence motifs, transcription factor expression, and CRE accessibility, we systematically reconstructed the gene regulatory networks that define cnidarian cell types. Our results reveal the regulatory complexity underlying cell differentiation in a morphologically simple animal and highlight conserved principles in animal gene regulation. This work provides a foundation for comparative regulatory genomics to understand the evolutionary emergence of animal cell type diversity. ### Competing Interest Statement The authors have declared no competing interest. European Research Council, https://ror.org/0472cxd90, ERC-StG 851647 Ministerio de Ciencia e Innovación, https://ror.org/05r0vyz12, PID2021-124757NB-I00, FPI Severo Ochoa PhD fellowship European Union, https://ror.org/019w4f821, Marie Skłodowska-Curie INTREPiD co-fund agreement 75442, Marie Skłodowska-Curie grant agreement 101031767
I am very happy to have posted my first bioRxiv preprint. A long time in the making - and still adding a few final touches to it - but we're excited to finally have it out there in the wild:
www.biorxiv.org/content/10.1...
Read below for a few highlights...
06.07.2025 18:14 — 👍 59 🔁 24 💬 1 📌 2
Fixed! Thanks for pointing it out
05.07.2025 19:02 — 👍 2 🔁 0 💬 0 📌 0
Enhorabuena to my first PhD sibling 🥰
02.07.2025 14:25 — 👍 13 🔁 0 💬 0 📌 0
🧬🔍How can enhancers achieve tissue-specific activity?
We use MPRAs of synthetic enhancers to derive interpretable rules on TFBS arrangement 🚦 and discover that negative synergies drive specificity in hematopoiesis 🩸. Shoutout to @Robert Frömel & @larsplus.bsky.social for leading this work 🦹🦸.
09.05.2025 06:39 — 👍 11 🔁 2 💬 0 📌 0

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
08.05.2025 16:06 — 👍 140 🔁 58 💬 4 📌 8

We released our preprint on the CREsted package. CREsted allows for complete modeling of cell type-specific enhancer codes from scATAC-seq data. We demonstrate CREsted’s robust functionality in various species and tissues, and in vivo validate our findings: www.biorxiv.org/content/10.1...
03.04.2025 14:30 — 👍 75 🔁 38 💬 1 📌 5
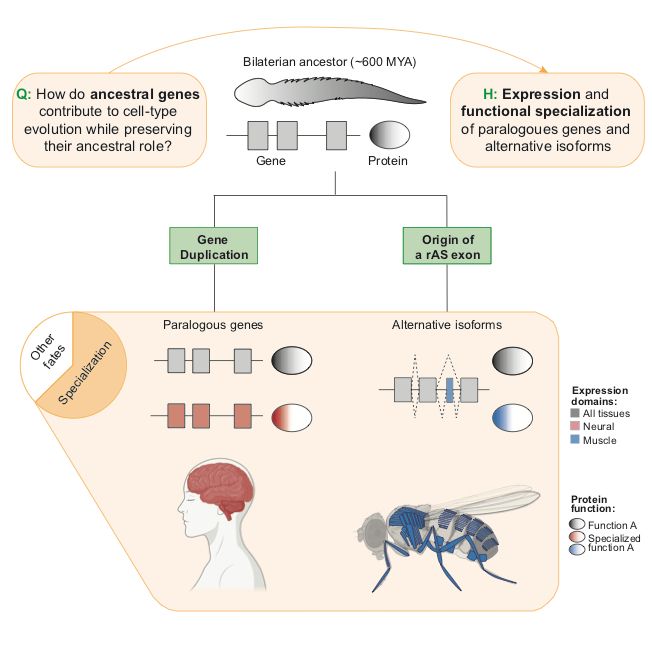
"The main fates after gene duplication are gene loss, redundancy, subfunctionalization and neofunctionalization".
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
18.03.2025 13:53 — 👍 41 🔁 16 💬 2 📌 1
EvoDevo & cnidarians.
Postdoc in the Sebé Pedrós lab at CRG. Previously PhD student in the Genikhovich group at Uni Vienna.
We are a lab focused on the study of invasive soil invertebrates, mainly land planarians. Led by Marta Álvarez Presas and based at the Universitat de Barcelona
Bioinformatics, genomics & gene regulation
Former PhD student at @zeitlingerlab.bsky.social
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Evolutionary Neuroscientist@Sussex, UK
PhD Neuroscience@University of Sussex w/ Tom Baden
MSc Neuroscience@University of Tübingen w/ Thomas Euler
Interested in visual neuroscience, neural design, evolution.
Assistant Prof. @ UMass Chan: studying gene regulation with multi-omic data and comp methods
www.moore-lab.org
Associate professor at the University of Barcelona.
Biologist
Scientist @arcinstitute
Ex–@stark_lab, @IMPvienna, @viennabiocenter
Scientist at IMP in Vienna. Excited about gene expression regulation and its encoding in our genomes - enhancers, transcription factors, co-factors, silencers, AI.
Senior research scientist at InstaDeep, interested in building computational models that can read the human genome and interpret it's variation
Postdoc @crg.eu Barcelona
GPCRs | synthetic biology | genomics
Doctoral candidate in the Uri Frank Lab. Studying epigenetics in Hydractinia.
Assistant Prof at Masaryk University. Dioscuri Centre for Stem Cell Biology and Metabolic Diseases #zebrafish #devbio 🇸🇰🇨🇿🇪🇺🇺🇸 , he/him, *All views my own
she/they. Master's student in Ecology and Ecosystems at the University of Vienna. In awe with the natural world and grateful to get to know it better.
(she/her) PhDing on jellyfish regeneration.
Finds tidbits of joy in random long walks and ice-creams!
Neuro and Developmental biologist. PostDoc at day. Supervillain at night. He/him #BlackLivesMatter #TransRightsAreHumanRights
@mads100tist@mastodon.social
Not-for-profit publisher of Development, Journal of Cell Science, Journal of Experimental Biology, Disease Models & Mechanisms and Biology Open. Host of community sites the Node, preLights and FocalPlane. Supporting biologists and inspiring biology.
The Evolutionary Medical Genomics (EvoMG) Program is a joint initiative by the CRG, UPF and IBE in Barcelona to boost biomedical research by applying evolutionary approaches.
Website: https://t.co/MBCtzyi8jt