Our nature note with @philtorres.bsky.social @aaronpomerantz.bsky.social et al. in
@ecol-evol.bsky.social was featured in @science.org #ScienceShots written by
@sahasmehra.bsky.social
#Cyclosa #decoy #spiders #evolution #Peru #Philippines
12.11.2025 23:28 —
👍 3
🔁 2
💬 0
📌 0
Congrats @philtorres.bsky.social and @aaronpomerantz.bsky.social on the paper! I look for these decoy spiders whenever I’m in the rainforest and this is the best example I’ve found. #inverts
Cuyabeno Reserve, Ecuador 2022
15.11.2025 12:22 —
👍 141
🔁 32
💬 3
📌 0
Hi Steven, here are a few that may be helpful -
The nanoMDBG study has some nice comparative datasets www.biorxiv.org/content/10.1...
Ultra-deep long-read metagenomics of soil microbes (used R9)
www.biorxiv.org/content/10.1...
Gut bacteriophage metagenomics pmc.ncbi.nlm.nih.gov/articles/PMC...
13.11.2025 07:00 —
👍 0
🔁 0
💬 0
📌 0

Yeast genomes show how large genomic variations affect trait diversity. #YeastGenomes #GenomicVariations #TraitDiversity #LongReads #Sequencing #Pangenome @nature.com 🧬 🖥️
www.nature.com/articles/d41...
18.10.2025 19:05 —
👍 1
🔁 2
💬 0
📌 0

Introducing our new Microbial Amplicon Barcoding Kit, which enhances microbial community profiling with an information-rich, rapid & accessible protocol for streamlined identification of bacteria, archaea & fungi — in one go. nanoporetech.com/news/oxford-...
18.09.2025 11:12 —
👍 13
🔁 4
💬 1
📌 0
🚨 New tool for viral genomics!
Meet CLAE: a high-fidelity Nanopore sequencing strategy that pushes accuracy to Q30, boosts throughput >800 Mb/100 pores, and recovers full-length viral genomes from complex samples 🌊🦠
👉 [https://doi.org/10.1002/advs.202505978]
#viromics #nanopore #genomics
12.09.2025 15:34 —
👍 17
🔁 8
💬 1
📌 0
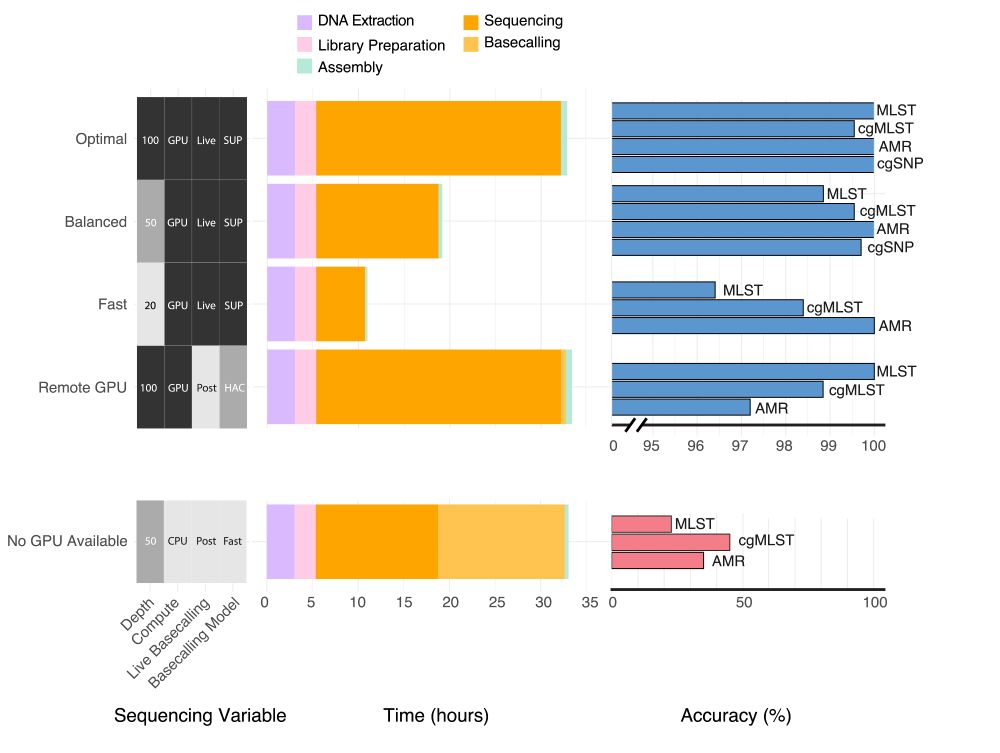
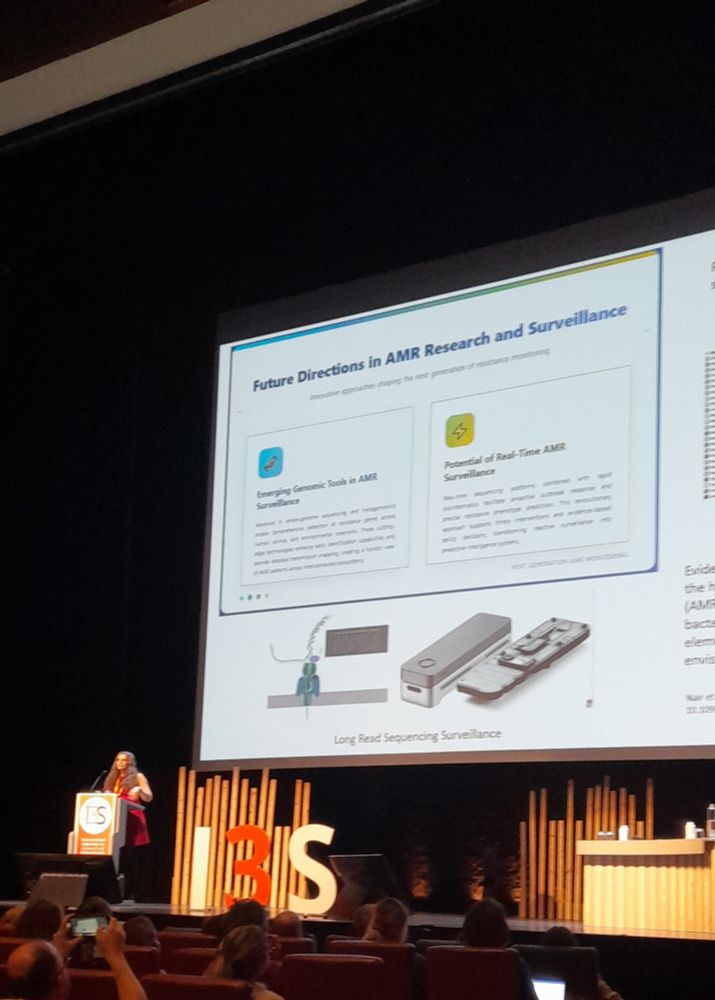
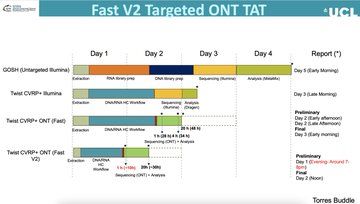
Pleased to say that our preprint benchmarking Nanopore data for MLST, cgMLST, cgSNP & AMR typing from bacterial isolates is out! TL;DR you can get almost perfect results from 50x depth using live SUP basecalling with a GPU in under 20 hours #microsky#IDsky 🦠🧬🖥️ /1
www.medrxiv.org/content/10.1...
30.07.2025 02:10 —
👍 46
🔁 29
💬 3
📌 2
Home | ATCC Genome Portal
The ATCC Genome Portal is a repository of high-quality, authenticated microbial reference genomes.
🧵🦠🖥️🧬🧪
Please RePost 🙏
We just released reference genomes for another 250 strains to the ATCC Genome Portal (genomes.ATCC.org)! We now have assemblies for
_5,750 microbial genomes_
AND
all assemblies w/ @nanoporetech.com data will include DNA methylation profiles too (details👇)
#genomics 1/
25.06.2025 20:06 —
👍 31
🔁 19
💬 1
📌 0

We’re bringing #ASMMicrobe 2025 to you, virtually.
Join Robert James, Angela Hickey, and Jose Alexander as they share how real-time, Oxford Nanopore sequencing is transforming microbiology—from single bacterial isolates to the complexities of the microbiome.
nanoporetech.com/about/events...
23.06.2025 13:01 —
👍 3
🔁 3
💬 2
📌 0
ATCC® 9945a™ | Bacillus paralicheniformis | ATCC Genome Portal
Bacillus paralicheniformis is a strain of bacteria in the Bacillus genus. This genome was published to the ATCC Genome Portal on 2022-04-02
Need bacterial DNA methylation data? Well, we’ve been re-basecalling all ONT data for the ATCC Genome Portal to get it. A huge effort, but as of today we have Mthyl data for ~1,500 bacterial strains. Attending #ASMicrobe? happy to chat IRL about it too. i.e. > genomes.atcc.org/genomes/67e8...
19.06.2025 20:39 —
👍 30
🔁 9
💬 2
📌 1
Francisella tularensis identified using @articnetwork.bsky.social SMART-9N metagenomics and @nanoporetech.com sequencing. Amazing work!
19.06.2025 10:27 —
👍 9
🔁 3
💬 1
📌 0
Very cool!
19.06.2025 21:59 —
👍 3
🔁 1
💬 1
📌 0
New metagenome assembler for ONT R10 and PacBio HiFi reads, from Jim Shaw. See Jim's thread for details
28.05.2025 19:07 —
👍 32
🔁 13
💬 0
📌 0
Short-read metagenomic sequencing cannot recover genomes from many abundant marine prokaryotes due to high strain heterogeneity and platform-inherent GC bias (likely viruses, too), but Nanopore long reads can address this. A results thread on our recent preprint 🧵.
25.05.2025 10:27 —
👍 44
🔁 23
💬 1
📌 4
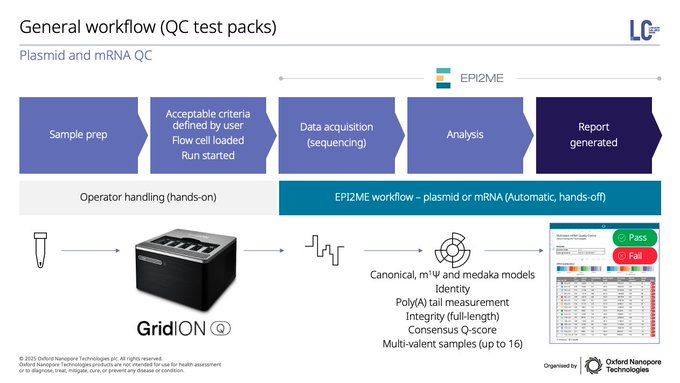
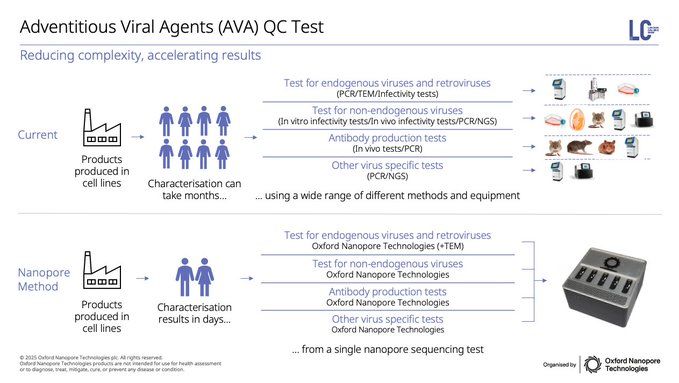
Last but certainly not least, we're taking these applications beyond research into applied industries & biopharma, including plasmid, AAV, mRNA manufacturing and adventitious viral agent contamination, provider deeper insight & faster TAT for these critical biological materials
23.05.2025 23:08 —
👍 0
🔁 0
💬 1
📌 0
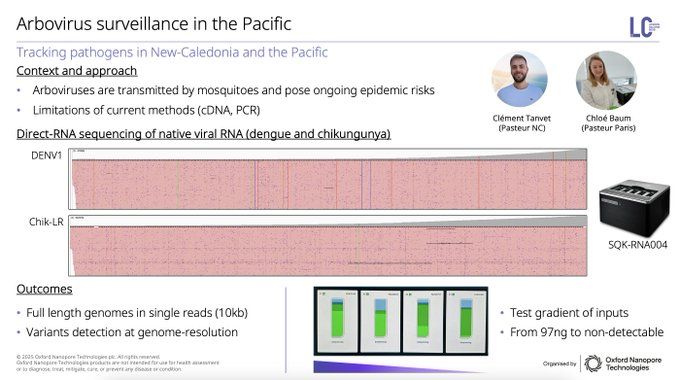
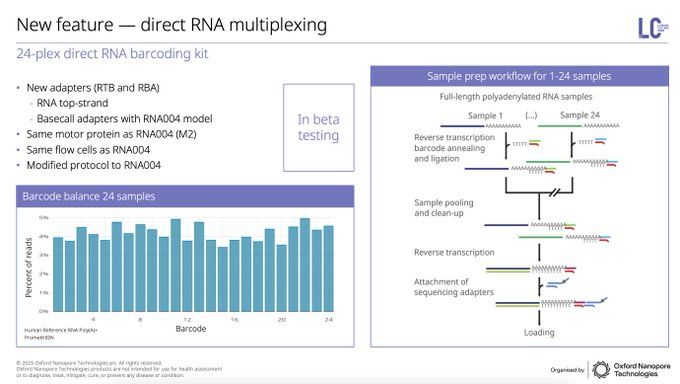
Direct RNA sequencing is also coming into the limelight for micro, highlighted by Chloé Baum at Pasteur Institute with dengue & chikungunya. Now with 24 barcodes coming for direct RNA multiplexing, this will become a critical tool for novel insights without biases of cDNA
23.05.2025 23:08 —
👍 0
🔁 0
💬 1
📌 0
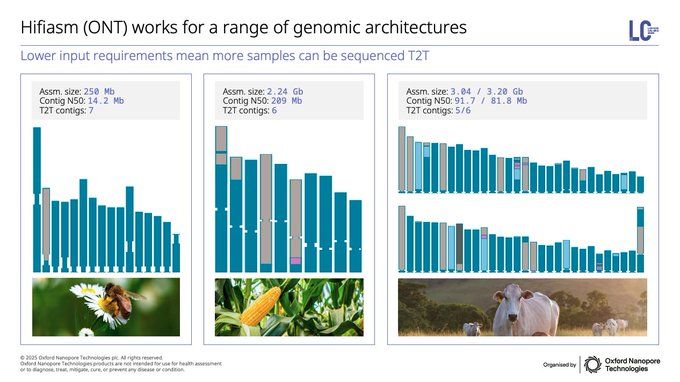
Bonus post sneaking in eukaryotes! High-quality, telomere-to-telomere (T2T) assemblies have potential to set a new gold standard, but were challenging & resource intensive. That's now changing thanks to new software like hifiasm-ONT - check out these amazing assemblies in 🐝🌽🐮
23.05.2025 23:08 —
👍 1
🔁 0
💬 1
📌 0
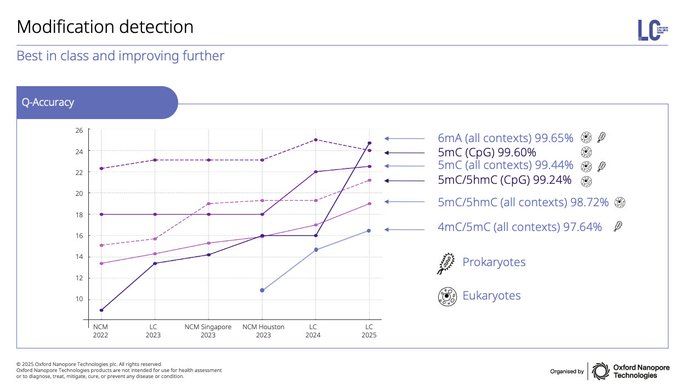
I also believe that the next frontier is Microbial Epigenomics. Detection of base modifications is best-in-class with @nanoporetech.com & the data is already right there every time you sequence a microbial sample directly without PCR. What discoveries are lurking right under our noses?
23.05.2025 23:08 —
👍 2
🔁 1
💬 1
📌 0
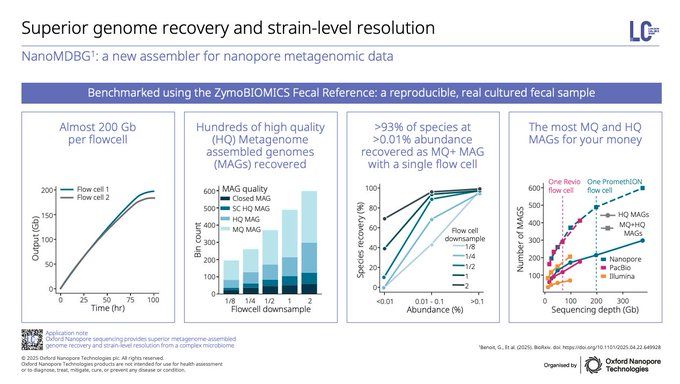
This extends beyond microbial isolates into complex microbiomes. @sisseljuul.bsky.social and @dorylophile.bsky.social showcased beautiful benchmarking data on superior genome recovery and strain-level resolution in the ZymoBIOMICS Fecal Reference with ONT compared to alternative sequencing platforms
23.05.2025 23:08 —
👍 1
🔁 1
💬 1
📌 0
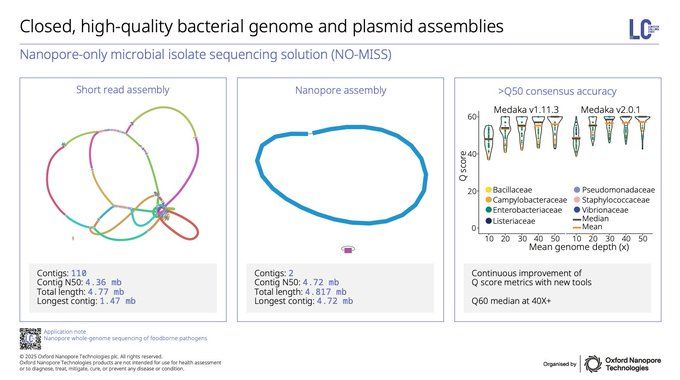
The ability to resolve closed microbial genomes & plasmids has been a critical case for long-reads, but our teams have been pushing to make ONT all you need - thanks to updates in basecalling/polishing, you now get best of both worlds: high-quality assemblies with high-accuracy
23.05.2025 23:08 —
👍 0
🔁 0
💬 1
📌 0
bsky.app/profile/nano...
23.05.2025 23:08 —
👍 0
🔁 0
💬 1
📌 0
bsky.app/profile/nano...
23.05.2025 23:08 —
👍 0
🔁 0
💬 1
📌 0