
Le CIRI recrute un Maître ou une Maîtresse de conférences en Microbiologie médicale
Plus d'informations : ciri.ens-lyon.fr/MCU-microbio...
@labxc.bsky.social
Group leader. HGT in bacterial pathogens. Acinetobacter, Legionella. Natural transformation. AMR. MGEs. Centre International de Recherche en Infectiologie

Le CIRI recrute un Maître ou une Maîtresse de conférences en Microbiologie médicale
Plus d'informations : ciri.ens-lyon.fr/MCU-microbio...
📢 Recrutement d’un(e) Maître(sses) de conférence en Biochimie générale, spécialité bactériologie moléculaire à MMSB
🔬Recherche
*Immunité anti-phages
*Réponse au stress, modulation de la croissance bactérienne
✉️ christophe.grangeasse@cnrs.fr
📚Enseignement : Université Lyon I
✉️ patrice.gouet@ibcp.fr
While YraN is likely guided by ComM to target the D-loop, CoiA which has replaced YraN in Bacillota, has evolved to specifically recognize the 3-strand junction.
Most bacteria carry one of these systems, one more evidence that natural transformation is a common feature in bacteria!

Without the nuclease/helicase, the rare recombination events are very short.
Hence, bacteria have evolved two distinct systems to efficiently resolve the D-loop and generate large recombination events by transformation
We analyzed hundreds of recombinaison events to understand how the two nuclease/helicase systems (YraN/ComM and CoiA/RadA) enable efficient transformation.
The defects in recombination in the absence of these very different nuclease/helicase systems are strinkingly similar!

Cleaving the displaced strand might be critical to allow for the helicase-driven extension of the D-loop. In vivo, genetic assays show that CoiA assists D-loop extension by the DnaB-type RadA helicase.
Similarly, YraN assists, and interacts with, the transformation-specific MCM-type ComM helicase

Indeed, in an in vitro-generated transformation D-loop, CoiA specifically cleaves the displaced strand at the edge of the D-loop
16.01.2026 16:23 — 👍 1 🔁 0 💬 1 📌 0
Could the nucleases be helping resolve the D-loop?
Biochemical analyses show that YraN is an endonuclease, cleaving ssDNA. CoiA is more specific, cleaving ssDNA if it's branching out of dsDNA.
Both proteins might cleaved the parental ssDNA displaced by the transforming DNA molecule

Our two labs separetely found that transformation requires a nuclease. CoiA in Streptococcus pneumoniae and a protein of unknown function in Legionella/Acinetobacter, YraN.
CoiA and YraN are different proteins of the PD-(D/E)XK family of phosphodiesterases.
Most bacteria possess one or the other

As they actively import DNA, bacteria process it into ssDNA. Then, RecA-dependent homology search leads the ssDNA to invade the chromosomal DNA, generating a 3-strand intermediate called a D-loop.
How this structure is resolved to stably integrate the transforming DNA is unknown
Since the discovery of natural transformation by Frederick Griffith in 1928, an ever increasing number of bacteria have been found to be able to integrate extracellular DNA it in their genome. This allows for acquisition/loss of genes and polymophisms, with consequence on #AMR
16.01.2026 16:23 — 👍 1 🔁 0 💬 1 📌 0#microsky
Massive update of preprint with @polardlab.bsky.social!
Bacteria have evolved two systems to recombine extracellular DNA
www.biorxiv.org/content/10.1...
Kudos to lead authors Léo Hardy, Violette Morales and Clothilde Rousseau, and to outstanding Dalia's lab and @epcrocha.bsky.social
🧵⬇️
We analyzed hundreds of recombinaison events to understand how the two nuclease/helicase systems (YraN/ComM and CoiA/RadA) enable efficient transformation.
The defects in recombination in the absence of these very different nuclease/helicase systems are strinkingly similar!

My team at @cbitoulouse.bsky.social is recruiting a postdoc #bioinformatics with solid experience in metagenomic analyses.
Interest in evolution, ecology & MGEs is important.
The offer stands until the perfect candidate is found, and it could be you 🫵
🔁 🙏
#microSky #phagesky #UTIsky
@cnrs.fr
Une opportunité unique de former des étudiants talentueux à la biologie des bactéries à l'ENS Lyon et mener des recherches sur les phages, l'immunité bactérienne, le transfert génétique horizontal (HGT) et/ou les éléments génétiques mobiles au sein du département de bactériologie du CIRI.
13.01.2026 14:01 — 👍 8 🔁 14 💬 0 📌 0Wonderful work! So satisfying to finally get a view of this critical step of natural transformation. Thank you for that.
Congratulations to all!

Pleased to share our recent article in PNAS - a collaboration with @jessicaandreani.bsky.social & Pablo Radicella, with important roles played by many members of each team.
A tripartite protein complex promotes DNA transport during natural transformation in Firmicutes www.pnas.org/doi/10.1073/...

🚨#PhD studentship opportunity! Plasmids provide bacteria with antimicrobial resistance, but do they have more fundamental effects on behaviour? 🧫🦠💫🧟♂️
Apply for a 4y funded MRC DiMeN position with me and Jamie Wheeler @livuni-ives.bsky.social www.findaphd.com/phds/project...

Our new paper is out in @narjournal.bsky.social We show that natural transformation enables bacteria to shuffle integron cassettes, boosting their phenotypic diversity.
academic.oup.com/nar/article/... 1/5

#ResultatScientifique 🔎 Certains gènes antiviraux humains partagent des défenses communes avec les bactéries, révélant une convergence évolutive surprenante 🧬
✍️ Alexandre Legrand et @lucievirevolte.bsky.social
📕 Nature Ecology and Evolution
buff.ly/Su0EdiU

#phahe #phagesky
www.biorxiv.org/content/10.1...
The ecology and evolution of bacterial immune systems'.
#phage #phagesky
A bunch of interesting reviews
royalsocietypublishing.org/doi/10.1098/...

Delighted to share our recently published work!
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇
What wonderful news! Congratulations, François, this is well-deserved recognition of your talent. I can't wait to see what you and your team will discover.
04.09.2025 14:02 — 👍 1 🔁 0 💬 0 📌 0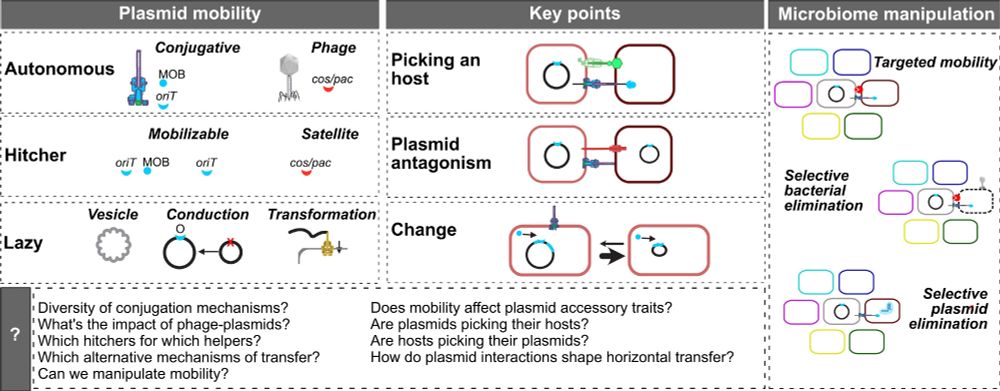
graphical abstract of the article the extended mobility of plasmids
Here's our new broad review on the extended mobility of plasmids, about all mechanisms driving and limiting their transfer. From conjugation to conduction, phage-plasmids to hitchers, molecular to evolutionary dynamics, ecology to biotech. The state of affairs. 1/9 academic.oup.com/nar/article/...
23.07.2025 07:35 — 👍 184 🔁 93 💬 4 📌 9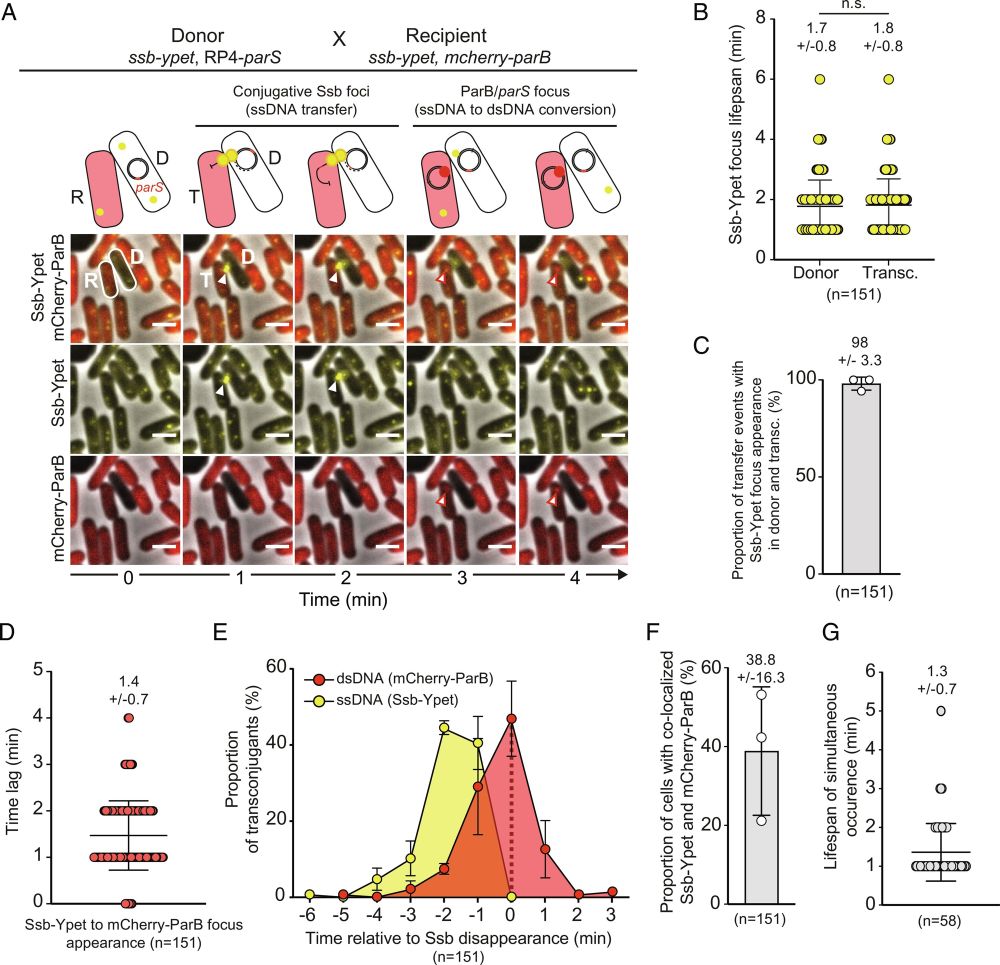
New publication from our Lesterlin lab in collab with lab of @knutdrescher.bsky.social
We determined the spatiotemporal dissemination of conjugative plasmids within biofilms and found that they spread only in specific structural regions.
www.pnas.org/doi/10.1073/...
I'm very happy to share the last paper from my previous postdoc with the amazing @epcrocha.bsky.social! Thanks to the reviewers for their feedback, which improved our work!
#Plasmids #AMR #HorizontalGeneTransfer #Evolution
Travel awards for trainees and early-stage investigators to attend the 2025 Acinetobacter conference are available. Deadline for applications is April 30. Please rt.
events.faseb.org/event/1b872d...

#TalentsCNRS 🥉 | Découvrez la vidéo de Lucie Etienne, chercheuse au Centre international de recherche en infectiologie et lauréate de la médaille de bronze du @cnrs.fr en 2024 !
@cnrsbiologie.bsky.social @lucievirevolte.bsky.social @ensdelyon.bsky.social @inserm.fr
www.youtube.com/watch?v=jE8b...
Frankly, I don't know how robust the data supported the probiotic role of Nissle 1917, but there is such large body of work on the genotoxic nature of colibactin and Nissle 1917 that I don't understand why Mutaflor has not yet been banned. Tagging @jpnoug.bsky.social
13.02.2025 14:51 — 👍 1 🔁 0 💬 2 📌 0