Schedule – Applied Systems Biology Symposia
Emmanuel Saliba, University of Würzburg, Germany.
Excited to present work from my preprint at #SMBI2025 @uni-jena.de on September 18th. I will be speaking on how spatial structure can explain the association of microbial accessory genes with inducible prophages.
asb-conference.hki-jena.de/schedule/
25.08.2025 18:17 — 👍 6 🔁 2 💬 0 📌 0

SpeSpeNet: an interactive and user-friendly tool to create and explore microbial correlation networks
Abstract. Correlation networks are commonly used to explore microbiome data. In these networks, nodes are taxa and edges represent correlations between the
📣 New, free webtool by the Snoek lab! Meet SpeSpeNet, an intuitive platform for automated network analysis of microbiomes to uncover complex interactions between microbes and their environments
@ismepublications.bsky.social @isme-microbes.bsky.social
doi.org/10.1093/isme...
#microsky #mevosky
14.03.2025 14:14 — 👍 11 🔁 9 💬 2 📌 0
Get a grasp of the chemistry of microbiomes! Check out Pilleriin Peets' preprint: she developed chemical characteristics vectors to reanalyze EMP's untargeted metabolomics data: www.biorxiv.org/content/10.1... with Aris Litos, Daniel Garza, Kai Dührkop, @jjjvanderhooft.bsky.social & Sebastian Böcker
27.01.2025 13:25 — 👍 11 🔁 3 💬 1 📌 0
We (with Clement Coclet, not on Bsky) had the chance to work on a broad "state of viromics" review. We tried to use this to give an overview of how the field changed over the last ~ 15 years, and also what we think are some of the major remaining challenges. Full-text access at -> rdcu.be/excHt
22.07.2025 15:06 — 👍 84 🔁 41 💬 2 📌 2

Examples of protein annotations across functional classification schemes. Yellow: hierarchical classification schemes, red: orthologous/homologous groups, and blue: methods using protein domain information. Protein structures were generated using the ColabFold Alphafold MMseqs2 interactive notebook (version 1.5.2) (57) and visualized using ChimeraX (58). (A) Functional labels associated with the glycogen synthase protein in Escherichia coli (UniprotKB P0A6U8). (B) Functional labels associated with Kehishuvirus sp. tikala major capsid protein (NCBI WEU69752.1).
New in @asm.org #MMBR Computational function prediction of bacteria and phage proteins. How to annotate your #phage and #bacteria genomes
by @susiegriggo.bsky.social @bedutilh.bsky.social @gbouras13.bsky.social and Bob
#phagesky #microsky
journals.asm.org/doi/epub/10....
22.08.2025 04:00 — 👍 51 🔁 27 💬 1 📌 0

PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning
AbstractBackground. Phages and plasmids are the major components of mobile genetic elements, and fragments from such elements generally co-exist with chrom
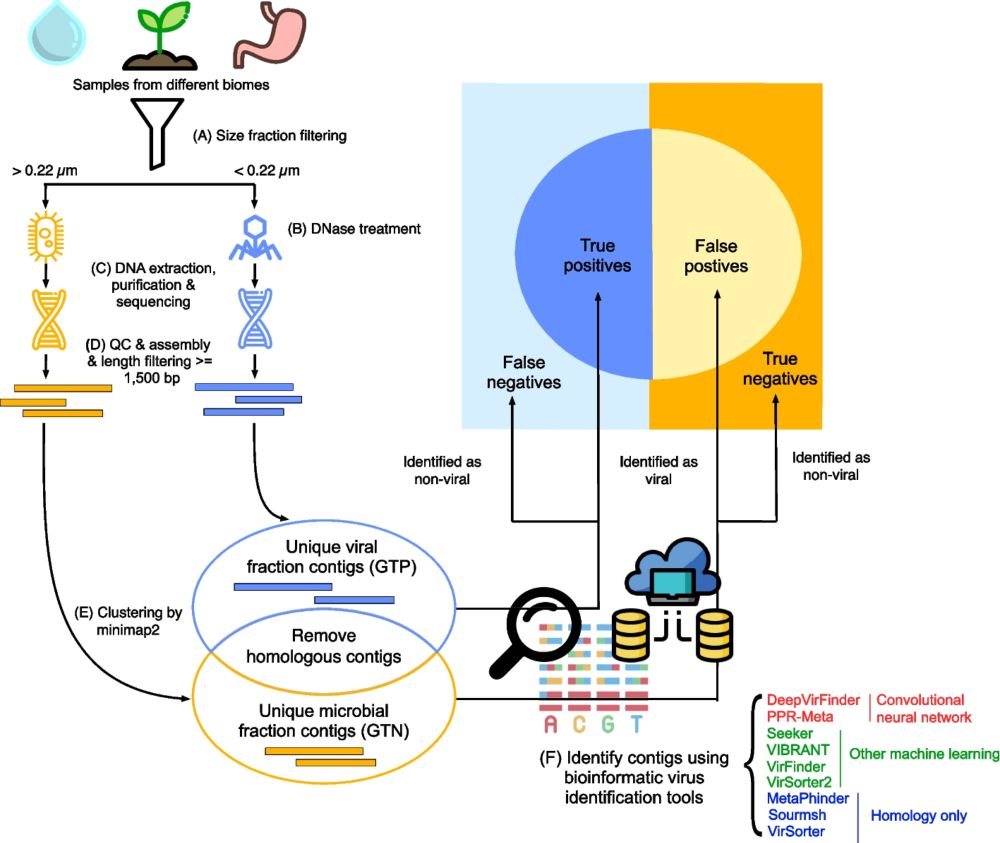
Tools' performance ranked similarly in different biomes. #MachineLearning 🤖 tools outperformed homology-based tools. #CNN tools performed the best. The best tool was PPR-Meta, which uses a CNN containing two paths—a “base path” and a “codon path”: academic.oup.com/gigascience/.... (3/4)
25.04.2024 08:30 — 👍 0 🔁 0 💬 0 📌 0
We used paired viral (TP) and microbial (TN) metagenomes from #Antarctic water 🇦🇶🌊, tomato soil 🍅🌱, and human gut 💩. Real-world data 1: has high complexity, 2: avoids training/testing overlap, and 3: is more realistic than cropping mock contigs from reference genomes. (2/4)
25.04.2024 08:29 — 👍 1 🔁 0 💬 0 📌 0
We are the largest biomedical science hub in the south of Europe, by the beach in Barcelona.
Recerca biomèdica amb vistes |
@researchmar.bsky.social @crg.eu @melisupf.bsky.social @embl.org @isglobal.org @ibe-barcelona.bsky.social @fpmaragall.bsky.social
Building genomic intelligence @ Tatta Bio
Prof Marine Virus Ecology
NIOZ - Royal Netherlands Institute for Sea Research
& Univ Amsterdam
The European Molecular Biology Laboratory drives visionary basic research and technology development in the life sciences. www.embl.org
don’t tell me you aren’t
✉️ you can also dm me your fav sci memes to post here
PhD candidate in Bioinformatics at Utrecht University
International Society for Viruses of Microorganisms (ISVM) aims to advance the science and use of viruses of microorganisms by supporting academic meetings, encouraging networking, and facilitating learning opportunities
Bringing together the excellence of virology and bioinformatics.
Tools: http://bit.ly/evbctools
Impressum: https://evbc.uni-jena.de/impressum/
Computational biologist | Bioinformatics, virus-host interactions, LLMs 🦠 💻
Joint-Postdoc in Küsel_lab, UniJena and Sullivan_lab, OhioState | alum UniGroningen| #firstgenphd | he/him/his 🏳️🌈| I study viruses
https://www.aadjiepratama.com
Microbial ecologist, postdoc at FSU
Biodiversity-loving, error bar-needing statistics nerd; Associate Professor @UWBiostat. Methods & software for #microbiome & #biodiversity data. She/her.
Bioinformatics @ University of Adelaide
- phages, microbes and more
Associate Professor
DFCI & HMS
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
PhD student at UChicago and the Marine Biological Laboratory studying bacteria and their associated viruses in coastal systems
Postdoc with @kayla-king.bsky.social and @sarperotto.bsky.social at UBC. Host-pathogen evolution. C. elegans, phages, and bacterial communities. Modeling and experimental evolution. Microbial and computational methods.
https://mikeblazanin.com/
Computational Biology PhD Student at the University of Birmingham. (Meta)genomic pathogen surveillance, infectious disease modelling, and BW disarmament. 🇬🇧/🇸🇬
Rutgers alumni, former post doc at Boston Children's hospital. Now studying bacterial stress response in Brazil.
#microbiology #Tuberculosis
Assistant professor at Wageningen University, interested in Comparative Microbial Genomics, Phage genomics, Evolution