
🥁 NEW
Our study on infant gut microbiome and #straintransmission is now published in @nature.com:
doi.org/10.1038/s415...
1/10

🥁 NEW
Our study on infant gut microbiome and #straintransmission is now published in @nature.com:
doi.org/10.1038/s415...
1/10

ACK (2/2) @cibiocm.bsky.social, all the participants of the PREDICT programme, Zoe Ltd, and all the funding agencies.
www.nature.com/articles/s41...
ACK (1/2) PaoloManghi @fackelmeister.bsky.social GabrielBaldanzi ElcoBakker @livi-ricci.bsky.social GianmarcoPiccinno ElisaPiperni KatarinaMladenovic FedericaAmati AlbertoArrè SajaysuryaGanesh FrancescaGiordano RichardDavies Jonathan Wolf KateMBermingham SarahEBerry TimDSpector NicolaSegata
10.12.2025 17:26 — 👍 1 🔁 1 💬 1 📌 0In conclusion, we provide a consistent and reproducible system for evaluating microbiome health conditions and support the direct, reproducible, and actionable link between diet and microbiome composition, which can be considered in future microbiome-powered precision nutrition research.
10.12.2025 17:26 — 👍 2 🔁 0 💬 1 📌 0
The two clinical trials consistently showed significant increases in species ranked favorably, while unfavorable species decreased over time, confirming that dietary changes can modulate the microbiome.
Of note, most of the favorably ranked species are previously uncharacterized or unknown.

We re-analyzed two independent dietary intervention trials: ZOE METHOD (NCT05273268, PDP; n = 177, and control; n = 170) and BIOME (NCT06231706, prebiotic; n = 116, control; n = 120, and probiotic; n = 113), and identified which species showed a significant change longitudinally in each arm.
10.12.2025 17:26 — 👍 1 🔁 0 💬 1 📌 0
These rankings were predictive for (i) BMI: individuals with healthy weight carried 5.2 more favorable SGBs on average than those with obesity; (ii) disease: healthy controls separated from cases (Type 2 Diabetes had a strong association with an unfavorable Cohen’s d score of −0.47).
10.12.2025 17:26 — 👍 0 🔁 0 💬 1 📌 0
We ranked 661 prevalent microbial species (most of which were unknown) based on their correlations with intermediate measures of cardiometabolic health (such as blood glucose, triglycerides, HDL, etc.) and diet (HEI, hPDI, uPDI, etc.) into two ranking systems.
10.12.2025 17:26 — 👍 0 🔁 0 💬 1 📌 0
I'm very happy to share our latest manuscript published today in @nature.com !!
➡️ doi.org/10.1038/s415...
We analyzed data from over 34,000 participants in the US and UK, revealing strong associations between specific gut bacteria and markers of cardiometabolic health and dietary habits.
🧬🖥️ Postdoc positions available SOON!!
Looking for researchers with strong computational skills (programming essential) to work on microbiome & phylogenetics projects.
📅 Start on Nov 1, 2025 (flexible)
📍 Trento, Italy
DM me for details or share!
#postdocjobs #microbiome #metagenomics #phylogenetics
Now @mireiavallesc.bsky.social presenting microbiome transmission at the #BCNmicrobiome
27.06.2025 09:53 — 👍 8 🔁 1 💬 0 📌 0
Heading to the Barcelona Debates on the Human Mocrobiome 2025 🇪🇸
I'll be talking about Microbiome-Diet and Cardiometabolic Health.
📍 CosmoCaixa
📆 June 26
⏰ 11:00
🔗 www.bcnmicrobiome.com
Come say hi if you're around! #bioinformatics #microbiome
Heading to Vienna this weekend! 🇦🇹
Thrilled to be talking about ML in bacterial genomics & #metagenomics at #ESCMIDGlobal 2025.
📍 Hall 4
📅 Monday (14/04)
⏰ 16:15 CEST
Come say hi if you're around! #bioinformatics #microbiome
cc @escmid.bsky.social
What I believe was surprising was that how much a single bacteria species is explaining (and predicting) a single food item (coffee), among all the complexities we have in microbiome studies. And these predictions were not presence/absence but rather with relative abundance values. (2/2)
21.02.2025 17:11 — 👍 3 🔁 0 💬 2 📌 2Thanks Gabriele for your interest in our work. Indeed, we don't really go into the clinical/medical relevance of Lawsonibacter here. (1/2)
21.02.2025 17:11 — 👍 3 🔁 0 💬 1 📌 0
MVIF january program
It's a new year!
...and a new #MVIF program is out! 🤩
Registration here: cassyni.com/s/mvif-35
⭐️MicroTalks:
Minna Chang
@hariszaf.bsky.social
⭐️ Keynote:
@chmoei.bsky.social
⭐️Selected talks:
Pratyay Sengupta
@fatimacpereira.bsky.social
@cladigar.bsky.social
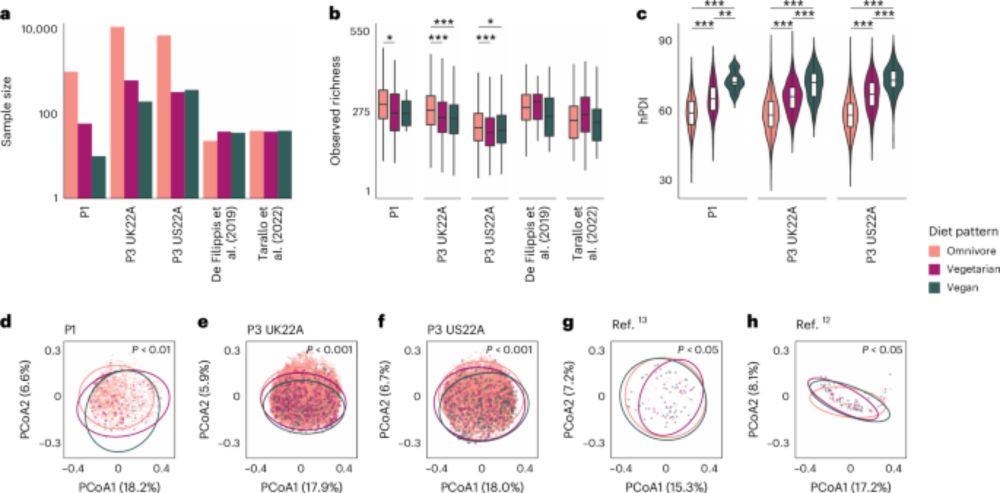
🧵 How does being a #vegan, #vegetarian or omnivore impact your gut #microbiome? 🥦 Find out in our new paper on gut microbiome signatures of vegan, vegetarian & omnivore diets & associated health outcomes across 21,561 individuals in @naturemicrobiol.bsky.social!🎉 1/10
www.nature.com/articles/s41...

Ciao @mikeneugent.bsky.social from the both us! (unfortunately mine is from the vending machine, the fastest one I could get here!)
17.12.2024 08:11 — 👍 3 🔁 1 💬 1 📌 0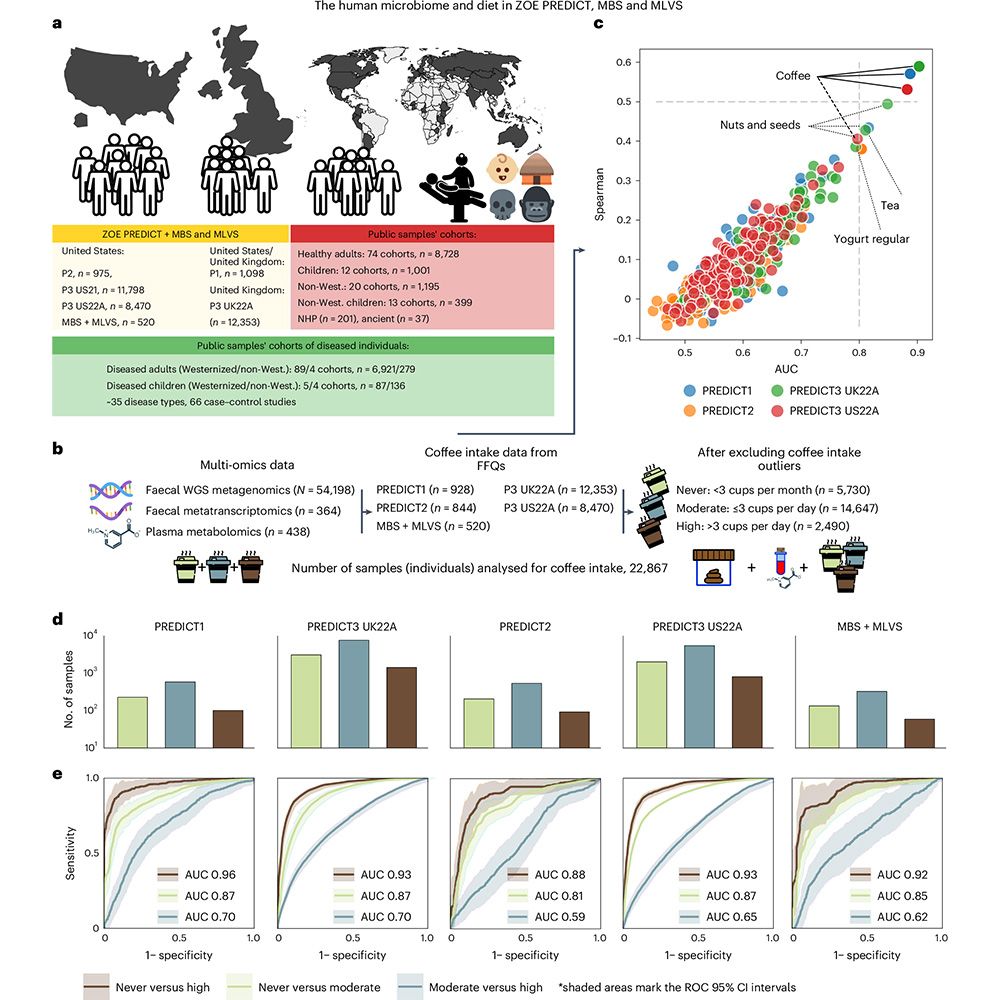
This is figure 1, which shows the consistent global links between coffee consumption and the human gut microbiome.
Coffee consumption is associated with the presence and abundance of a specific member of the human gut microbiome, Lawsonibacter asaccharolyticus, and changes to the plasma metabolome, according to a paper in Nature Microbiology. https://go.nature.com/3CIl4as 🧪
24.11.2024 16:08 — 👍 140 🔁 31 💬 4 📌 6



Fantastic event in Goiania! Beautiful audience and nice people! Had a great time, thanks Joao for the invitation and all the organizers for the organization!
24.11.2024 14:33 — 👍 2 🔁 0 💬 0 📌 0
Thrilled to speak at the "I Brazilian Conference on Nutrition, Gut Microbiome, Health, and Disease", looking forward to meet #microbiome scientists in Brazil! www.even3.com.br/i-brazilian-...
22.11.2024 06:48 — 👍 10 🔁 1 💬 1 📌 0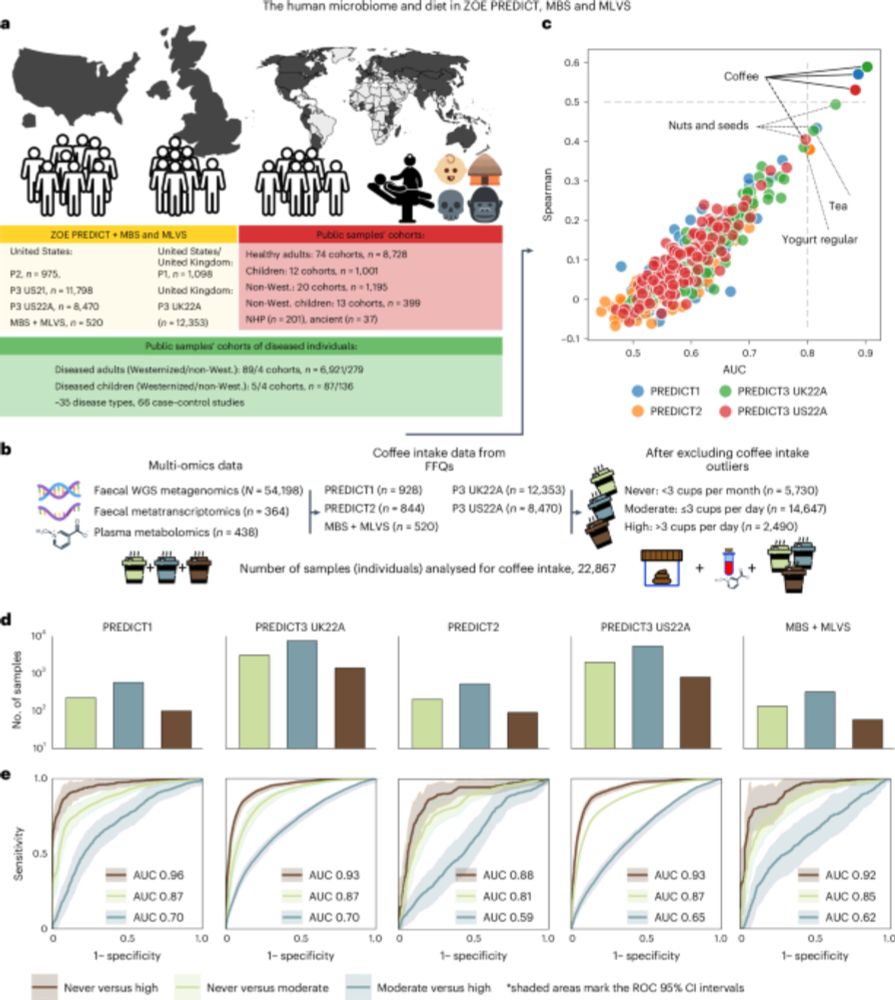
🥁 New Publication Alert:
Our manuscript on the ☕️ and #microbiome relationship is out in #NatureMicrobiology:
www.nature.com/articles/s41...
1/12
Would love to be included if possible, thanks a lot
18.11.2024 09:05 — 👍 1 🔁 0 💬 0 📌 0
Be aware of common pitfalls when using ML in microbiological studies (Box 2) and hopefully, the simplified checklist when reading, reviewing, or applying ML (Box 3) can help in better understanding the best strategies to use and their evaluation.
6/6
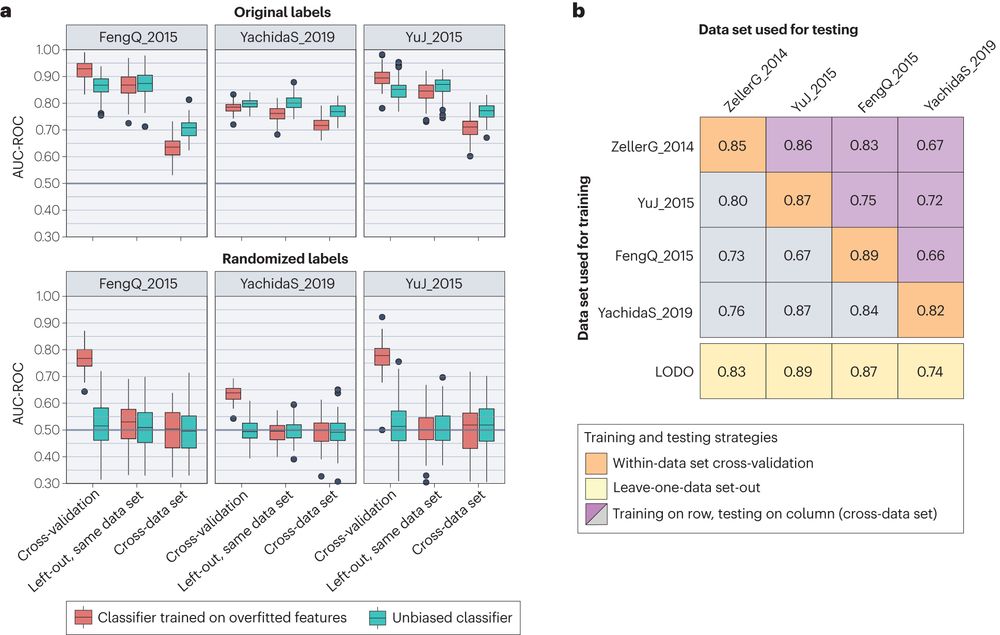
Be careful of overfitting, feature selection and testing within the same data, which could lead to overoptimistic within-dataset and poor performances cross-cohorts.
5/6
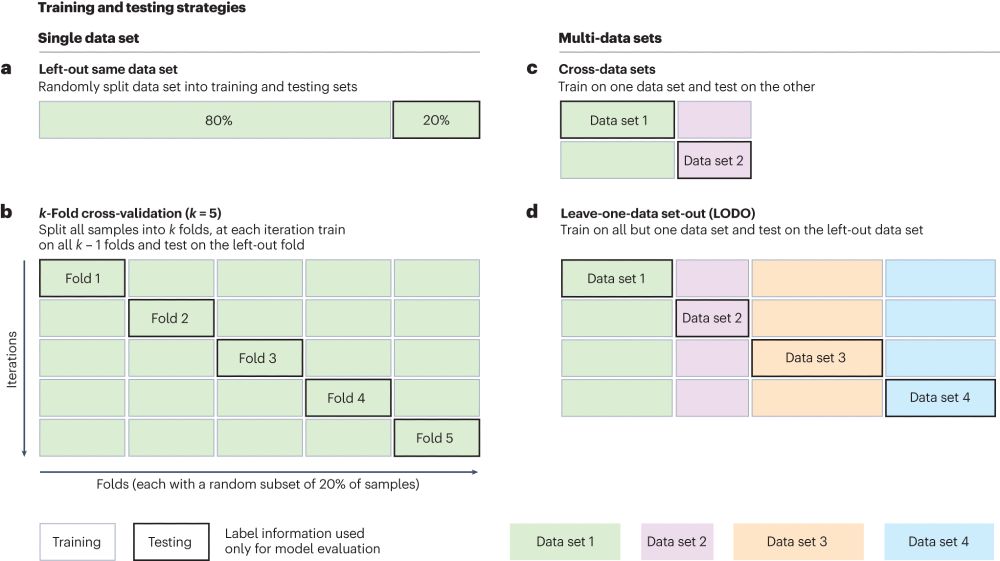
Several strategies are available to accurately estimate ML performances and depend and the data/cohorts availability.
4/6
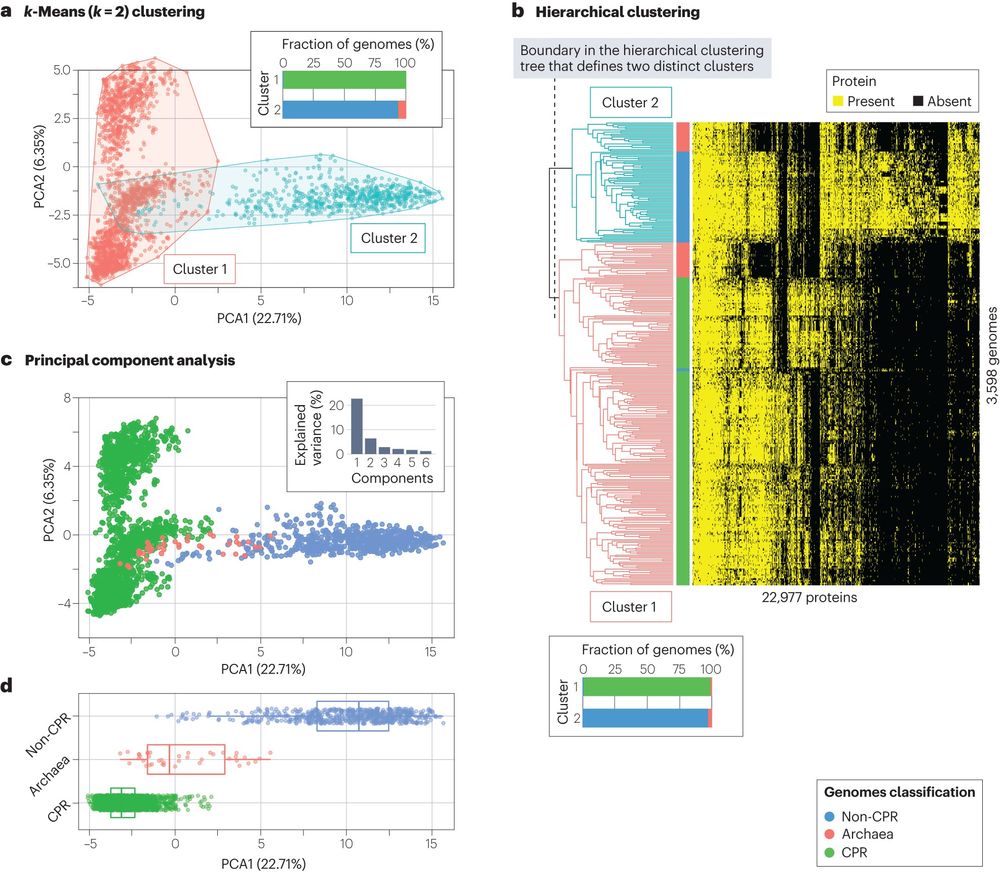
Dimensionality reduction and clustering are two of the most used algorithms in microbiology, and different algorithms will have different results and visualization of high-dimensional data is also a non-easy task.
3/6
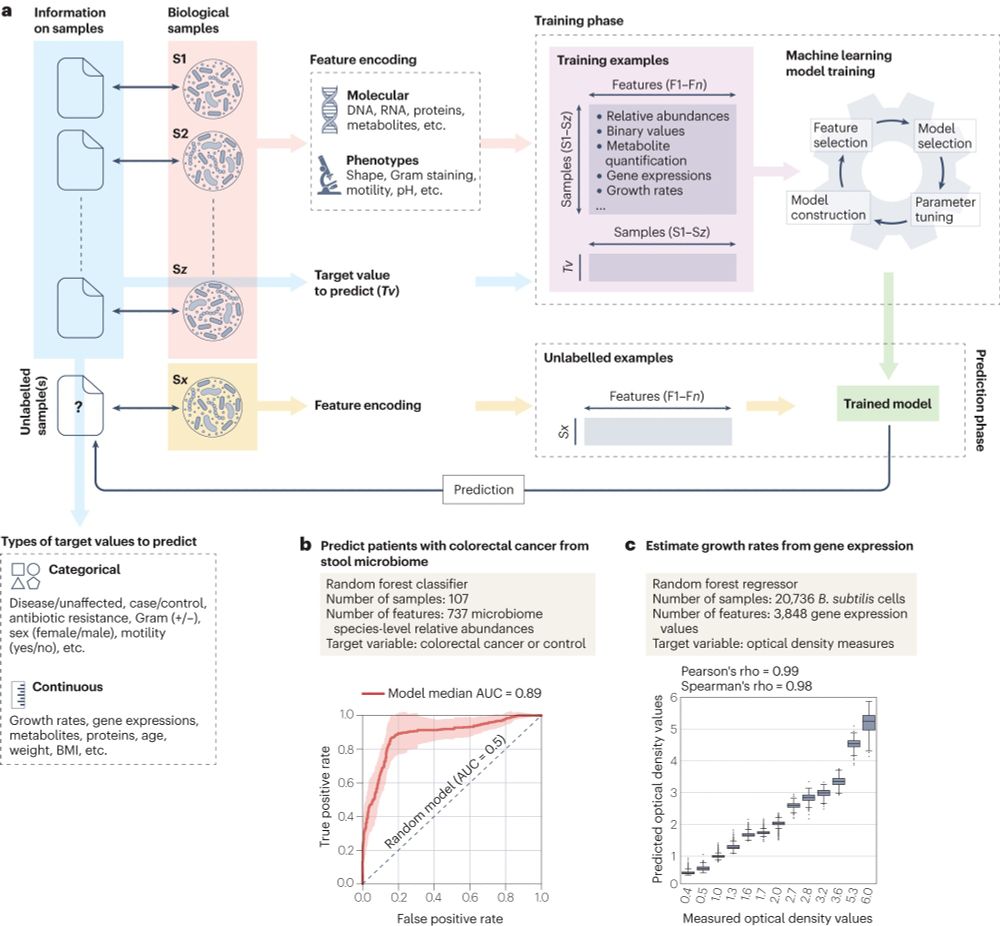
A ML workflow involves many small details: difficult to represent in a simplified schema as most are also algorithm-dependent. However, having a clear idea about the process can better help understand how several steps (samples, features, training, prediction) connect.
2/6
I'm thrilled to share our team's effort towards enabling ML usage and evaluation for microbiologists. Here goes the summary thread 🧵
1/6