Excited and proud to share our new work with @cibiocm.bsky.social, @vheidrich.bsky.social and @michal puncochar!
21.01.2026 18:31 — 👍 3 🔁 3 💬 0 📌 0Excited and proud to share our new work with @cibiocm.bsky.social, @vheidrich.bsky.social and @michal puncochar!
21.01.2026 18:31 — 👍 3 🔁 3 💬 0 📌 0Paolo Ghensi, V Heidrich, D Bazzani, F Asnicar (@fasnicar.bsky.social), F Armanini, A Bertelle, F Dell'Acqua, E Dellasega, R Waldner, D Vicentini, M Bolzan, L Trevisiol, C Tomasi, E Pasolli (@epasolli.bsky.social), Nicola Segata
05.06.2025 09:55 — 👍 1 🔁 0 💬 0 📌 0
We also used machine learning to show that diseased implants could be identified from microbiome profiles alone, reinforcing the potential of assessing the peri-implant microbiome in the clinical setting as a supportive diagnostic tool
05.06.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0
These included many functional and taxonomic unknowns, but also the well-studied for its role in periodontitis Fusobacterium nucleatum, with specific F. nuc clades associated differentially with mucositis and peri-implantitis
05.06.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0
Here we expanded our previous cohort (Ghensi et al. 2020) and used improved metagenomic profiling tools to identify strong microbiome markers of implant health and disease (mucositis or peri-implantitis)
05.06.2025 09:55 — 👍 0 🔁 0 💬 1 📌 0
In this study just published at the Journal of Clinical Periodontology - a collaboration between @cibiocm.bsky.social and PreBiomics - we evaluated the microbiome of dental implants using metagenomic sequencing
onlinelibrary.wiley.com/doi/10.1111/...
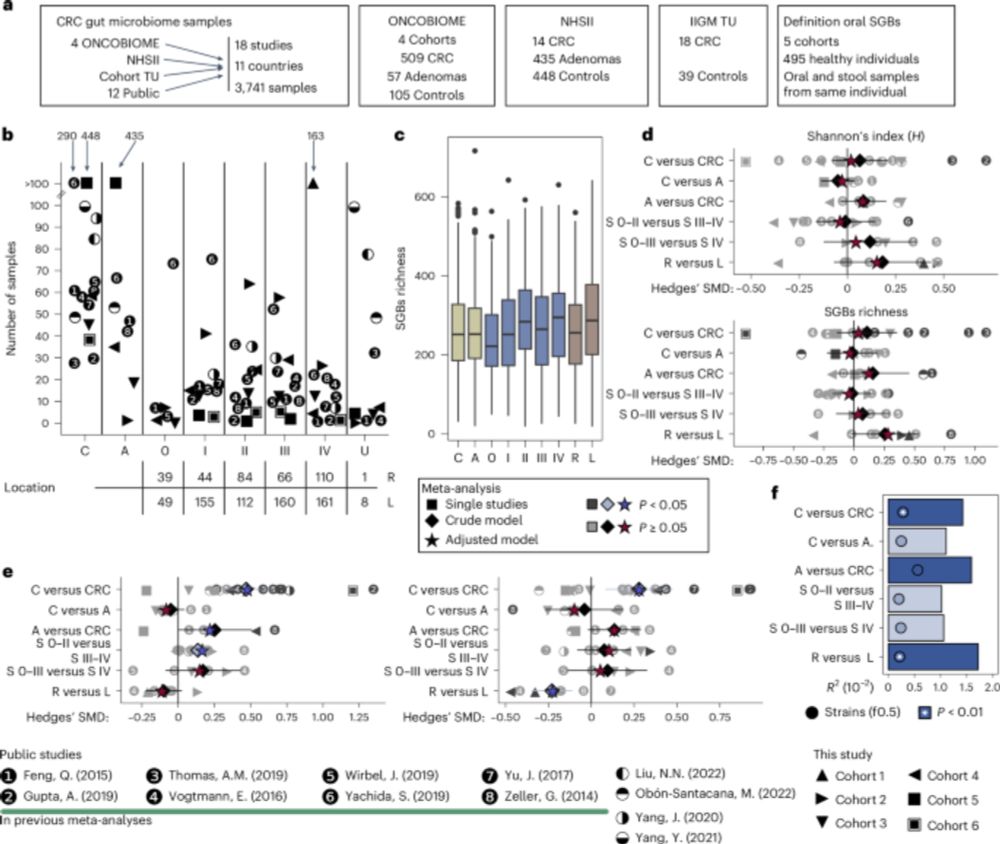
🥁NEW publication from our lab:
the largest meta-analysis of gut microbiome associations with CRC!
www.nature.com/articles/s41...
🧵⬇️
1/6

Thrilled to share that our manuscript on strain-level gut microbiome variation across diverse populations and human phenotypes is out today in @cellpress.bsky.social
Curious about how strain diversity relates to human traits? Follow this thread! 🌍 (1/n)
www.sciencedirect.com/science/arti...
Full-text: t.co/G8zk3j9Vtm
22.03.2025 19:03 — 👍 0 🔁 0 💬 0 📌 0
Our review on the acquisition and transmission of the human microbiome, now out @naturerevmicro.bsky.social 🦠 🔄
This was a fun writing process with @vheidrich.bsky.social and Nicola Segata! See Vitor's summary thread below:

Finally, we discuss the impact that microbiome transmission can have on human health and disease, and how it is possible to leverage this knowledge for the design of effective microbiome-targeting strategies
www.nature.com/articles/s41...
Have a nice read!
@cibiocm.bsky.social

We also list the social, host-related, and microbial factors potentially associated with human microbiome transmission, and talk about the methodological challenges associated with microbiome transmission inference
21.03.2025 16:02 — 👍 1 🔁 0 💬 1 📌 0
Since this includes microbial exchange not only with the main fount of our microbes - other humans, we dissect all the potential sources of human microbiome strains, including animals and foods
21.03.2025 16:02 — 👍 1 🔁 0 💬 1 📌 0We discuss how the building blocks of our microbiome - single microbial strains - are dynamically exchanged with the surroundings, leading to the continued acquisition and transmission of microbiome members
21.03.2025 16:02 — 👍 0 🔁 0 💬 1 📌 0
Thrilled to share our review (with @mireiavallesc.bsky.social and Nicola Segata) on "Human microbiome acquisition and transmission" just published @naturerevmicro.bsky.social
21.03.2025 16:02 — 👍 36 🔁 17 💬 1 📌 3
🥁
We are happy to announce the release of a new version of curatedFoodMetagenomicData (cFMD)
🔗 Get it here: github.com/SegataLab/cFMD
🧵👇
1/5

Wrong figure here, right one below
18.02.2025 17:16 — 👍 0 🔁 0 💬 0 📌 0
Link to the publication: rdcu.be/eajx6
@fackelmeister.bsky.social @raj09.bsky.social @cengoni.bsky.social @cibiocm.bsky.social
In conclusion, we hope our results and the better ability to metagenomically profile oral samples from dogs enabled by our study will encourage further efforts to characterize the microbiomes of pets (7/7)
18.02.2025 09:16 — 👍 0 🔁 1 💬 1 📌 0This finding provides some support for the continued use of dogs as models of periodontal conditions in humans and, more interestingly, hints at the possibility of the acquisition of oral pathogens from companion animals (6/7)
18.02.2025 09:16 — 👍 0 🔁 1 💬 1 📌 0This is probably linked to the very different anatomy, environmental conditions, and hygiene levels found in each host species' oral cavity. Despite this limited overlap, we noticed that species found in both hosts were often periodontal pathobionts, such as Campylobacter rectus (5/7)
18.02.2025 09:16 — 👍 1 🔁 1 💬 1 📌 0
We found the dog plaque microbiome is more diverse than the human plaque microbiome and that there is very little microbial overlap between the dental plaque of dogs and humans (only ~6% of species found in both host species) (4/7)
18.02.2025 09:16 — 👍 1 🔁 1 💬 1 📌 0
We then added these new dog plaque species to the database of MetaPhlAn 4 (github.com/biobakery/Me...) to be able to quantify their abundance in metagenomes. This allowed us to evaluate the overlap between the plaque microbiome of dogs and humans (3/7)
18.02.2025 09:16 — 👍 2 🔁 0 💬 1 📌 0
Through metagenomic sequencing and assembly, we reconstructed genomes for 347 dog plaque microbial species (spanning one archaeal and 17 bacterial phyla), 265 of which had never been identified before (2/7)
18.02.2025 09:16 — 👍 1 🔁 1 💬 2 📌 0
In this study just published at npj Biofilms and Microbiomes, we collected dental swab samples from dogs to shed light on the hardly studied dog dental plaque microbiome (1/7)
18.02.2025 09:16 — 👍 5 🔁 2 💬 2 📌 1