#HelloQuitteX #20Janvier
20.01.2025 13:53 — 👍 3 🔁 0 💬 0 📌 0
#syntheticbiology #synbio #devbio #axialelongation #morphogenesis #preprint
17.12.2024 14:06 — 👍 1 🔁 0 💬 0 📌 0
@steffengrosser.bsky.social, who performed tissue fluidity analysis.
and Calvin Lam, who performed, ages ago, the first simulations of a potential circuit for tissue elongation!
17.12.2024 14:03 — 👍 2 🔁 0 💬 1 📌 1
Thanks to the whole team!
@leonardomorsut.bsky.social, our beloved PI,
Christian Chung, Naisargee Jain, Catcher Salazar: wonderful students I had the pleasure to manage for this project, their work was critical!
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
This concludes this "skytorial" ("blueditorial"?), we are looking forward to reviewers comments, and yours!
Any feedback will help us reach this milestone of synthetic developmental engineering: the programming of any tissue shapes through guided self-organization.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
We need better cell engineering methods for inserting larger payloads with many genes induced by the same transcription factor.
And/or methods to avoid gene silencing, so we can do the same in a stepwise manner without losing the induction of the fist gene by the time we add the third!
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
For the 2nd and 3rd goals, we think the way is to screen more effectors, alone and in combination.
We tried to, but were faced with the common bane of synthetic biologists: gene silencing!
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

For the 1st goal, we have an idea of how to proceed:
Have the OFF transceivers express P-cad, and the ON transceiver N-cad.
This just requires a NOT gate downstream of synNotch.
...We have the plasmids already cloned, now we just need a brave student or postdoc to use them!
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
What we learn from this visualization method is that we need to:
1st: improve segregation of the “growing tip” and “structural support” regions,
2nd: further decrease tissue fluidity.
3rd: further control tissue growth speed.
... still a lot of work!
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

We then simulated elongation circuits in this “realistically bound” parameter space.
To better understand how our in vitro results compared to in silico ones, we used a visualization approach with a “morphospace” framework, inspired by @ricardsole.bsky.social
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
At this point, we have obtained the first elongating phenotype guided by synthetic gene circuits…
But we were still far of what the computational model promised… Probably in part because of its incomplete parametrization.
=> We re-parametrized the model with all our growth and fluidity data.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

In the second class, we induced N-cad (+/- p21) in activated transceivers, and this too led to tissue elongation.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
We combined our findings into 2 classes of “complete” circuits.
In the first class, we did not induce N-cad but had it constitutively expressed (its induction increased growth...)
And, lo and behold! When inducing the most promising fluidity control effectors, we obtained elongating structures!
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

At this point, we implemented 2/3 target design principles.
So, what about enforcing the segregation of the “growing tip” region?
=> We found that inducing N-cad upon transceivers activation (so, in the “structural support” region) would lead to better segregation! (We are not sure why)
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

Coming back to our design: we then screened candidate effectors for controlling tissue fluidity.
Here, we believe we share the first screen of this type, as we had to look for inspiration in many publications!
Our two best hits targeted actomyosin contractility: constitutively active RHOA & MLCK.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
Side note: there still is a need for better and more specific genes controlling tissue growth.
We probably need to:
1) screen a large amount of (mutant) genes (directed evolution?)
2) simultaneously induce multiple effectors. We have prelim. data showing this works, the limit is cell engineering
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

… The most efficient growth inhibitors also increased tissue rounding speed!
(We do not yet know why.)
p21 was the only effector which both decreased tissue growth and fluidity, so we decided to build upon its induction.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

With this method, we first screened effectors to inhibit tissue growth.
We found some promising ones, like p53.
… but there was a hidden issue with those…
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
We then screened effectors that could control tissue physics (growth and fluidity)
To test them in the most relevant context, we devised a pipeline where their impact could be evaluated in the right context: under the control of the transceiver circuit itself.
For more details, check Supp Fig 4 😉
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

We first identified a transceiver clone with a fate propagation speed in the right range. (the "Fast" clone here)
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
Now that we had a proof of concept in a computational model, we decided to go forward and implement this in real cells (mammalian fibroblasts: L929).
As we were trying to build a complicated circuit, we decided to proceed in a stepwise manner, splitting our work in semi-independent modules.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
We used this model to identify 3 important design principles:
1) strong difference in growth rate between the “growing tip” and “structural support” regions.
2) strong segregation of the “growing tip”, through the right adhesion matrix
3) high rigidity of the “structural support” region.
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
To verify if this would work, we first implemented the circuit in our own CompuCell3D framework: pubs.acs.org/doi/abs/10.1021/acssynbio.0c00369
… and this worked great! (...before physics were completely parametrized!)
(color code mistake here, the growing tip is blue instead of gray!)
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

So, how to exploit this circuit design?
We got the idea that if transceivers changed their properties based on their activation status, this could result in tissue elongation.
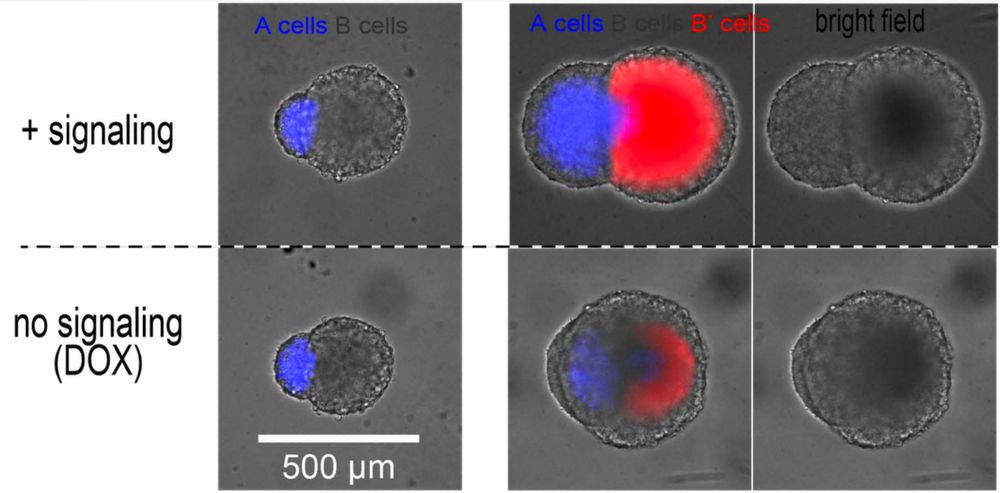
For that, the cells must only proliferate when OFF (grey), and become collectively very rigid when ON (red).
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
This circuit can be used to propagate a change in cell fate in 2D in a wave-like pattern… And also in 3D!
In this animation, a spheroid of “sender” cells (red+green=yellow) is fused with a spheroid of “transceiver” cells. Activated transceivers become green.
17.12.2024 14:03 — 👍 12 🔁 2 💬 1 📌 1

But, how to translate this concept to a synthetic gene circuit?
We were very lucky… We could just take advantage of another circuit we developed in the lab, the “transceiver” circuit!
(here: www.nature.com/articles/s41467-024-53078-8)
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

Looking at other (very different!) examples, one can notice a common design principle:
A combination of local material addition (to the tip) with mechanical resistance to rounding forces (of the bulk).
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0

So, how did we do that?
First, of course, we got our initial inspiration from the literature on axial elongation.
Notably this wonderful 2018 publication from @campaslab.bsky.social where the team demonstrated how tissue fluidity supports axial elongation in the zebrafish embryo!
17.12.2024 14:03 — 👍 1 🔁 0 💬 1 📌 0

While we were aiming to generate baguettes, we obtained something which is more in the range of olives / cocktail sausages, but, well, you have to start somewhere!
Here is a 20-ish posts summary, check my repost of Leonardo’s recap for a shorter version 😉
17.12.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0
Delighted to share our last preprint!!! 🎉🎉🎉
biorxiv.org/content/10.1101/2024.12.11.627621v1
We share the first synthetic gene circuit guiding the self-organization of mammalian tissues in elongating structures by dynamically tuning growth, viscosity and adhesion.
17.12.2024 14:03 — 👍 18 🔁 8 💬 2 📌 1
Scientist + Teacher UvA/Amsterdam | Cells | Molecules | Microscopy Quantitative imaging | Fluorescent Proteins | Biosensors | Open Science | dataViz | R | web apps
Homepage: https://joachimgoedhart.github.io/
DataViz Apps: https://huygens.science.uva.nl
Stem cell biologist wanting build an artificial kidney and understand development. My lab is at https://lindstromlab.usc.edu/
Director of ASCEND
See our spatial transcriptomic, organoid, and stem cell program https://ascend.usc.edu
Our goal is to turn you from someone who wants to play TRPGs into someone who wants to run them.
Cilia and cell motility enthusiast, basal cognition, weird organisms esp protists and larvae, how do living systems compute?
Professor of Cellular & Biophysical Dynamics, Living Systems Institute, Exeter (past: DAMTP, Cambridge)
www.micromotility.com
Scientist, mother of 3, art lover, physics of life, (bottom-up) synthetic biology, evolution, philosophy of science, TU Delft, www.tudelft.nl/laanlab, founder BiBB https://www.betainbestuurenbeleid.nl/nl/about-us
We combine soft matter physics, biophysics and cell biology to uncover physical principles of cellular organization @ Cluster of Excellence Physics of Life, Dresden
Curiosity driven science in physics of life and frugal innovations for planetary scale challenges
https://bhamla.gatech.edu/
Group leader at EMBL Heidelberg, previously: Postdoc at Yale with Ben Machta, PhD at LMU Munich with Erwin Frey; Theoretical Physicist by training & interested in Physics of Life
TLM is an initiative to build a network for researchers interested in theory and modelling of biological phenomena. We are on X (old twitter) @TLM_Cambridge, @TLM_Cambridge@mstdn.science. Or follow our website for more details: tlmcambridge.blog
Interested in protein self-organization. We rebuild the bacterial cell division machinery and small GTPase networks in vitro. https://looselab.ist.ac.at
Associate Professor
MIT Physics
Group Leader @MPI-CBG and @POL, TU-Dresden. Dev Biologist mixing it up w/ Physics. Want to know how organs grow! also obsessed with structural colors | ritamateus.com
Theoretical biophysics group at MPI-DS, Göttingen. We study the spatiotemporal organization of soft matter in cells, tissues, and synthetic systems; see www.zwickergroup.org
Theoretical Physicist studying the dynamics of living systems across scales, Associate Professor @ Georgia Tech
How do organs form from cells to tissue? Zebrafish and organoids; live imaging; quantitative biology; theory. Comments by Caren Norden
Physical principles of regeneration in zebrafish. Assist. Prof. @Genevunige. Passionate of zebrafish scales, bone morphogenesis and signalling dynamics. #SciComm and Improv theatre. https://tinyurl.com/23str3yn
Professor at Warwick Medical School interested in how complex shape emerges during embryo development. Will work on anything that can be imaged live :)
Professor of Developmental Biophysics.
Crossing all disciplines, through science, art and food | UCL | LMCB | IPLS
www.tissuemechanicslab.com
Science communication and #SciArt via www.datascaperealities.com
Loves hiking, glamping and sourdough baking















