
Our most recent publication with our collaborators at Novartis Biomedical Research uses the idea of Cooperative Free Energy in order to understand the thermodynamics of cooperativity in ternary complexes.
pubs.acs.org/doi/10.1021/...
@rinikerlab.bsky.social
Riniker research group, ETH Zurich

Our most recent publication with our collaborators at Novartis Biomedical Research uses the idea of Cooperative Free Energy in order to understand the thermodynamics of cooperativity in ternary complexes.
pubs.acs.org/doi/10.1021/...
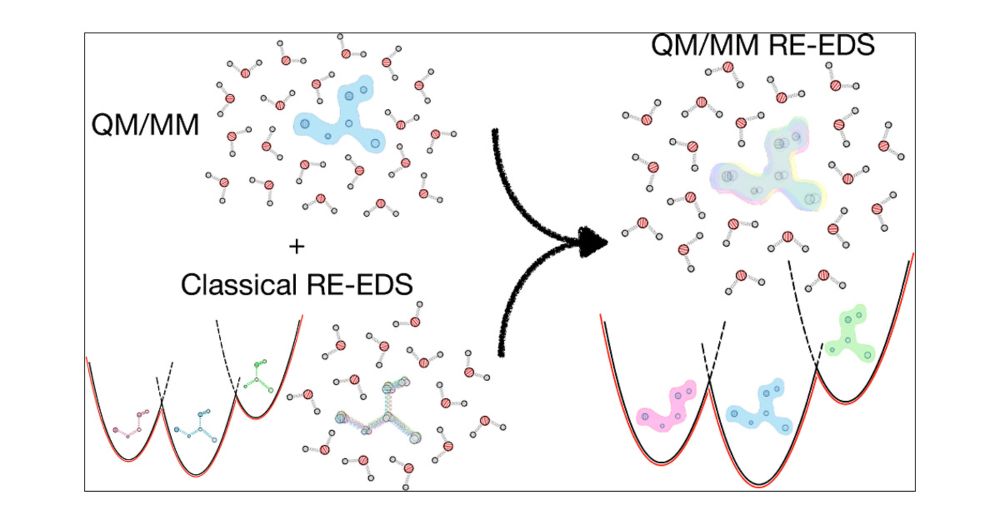
Our newest publication explores using QM/MM together with RE-EDS to do efficient free-energy calculations, enabling increased accuracy and allowing highly polarized systems to be studied.
#chemsky #compchem
doi.org/10.1021/acs....
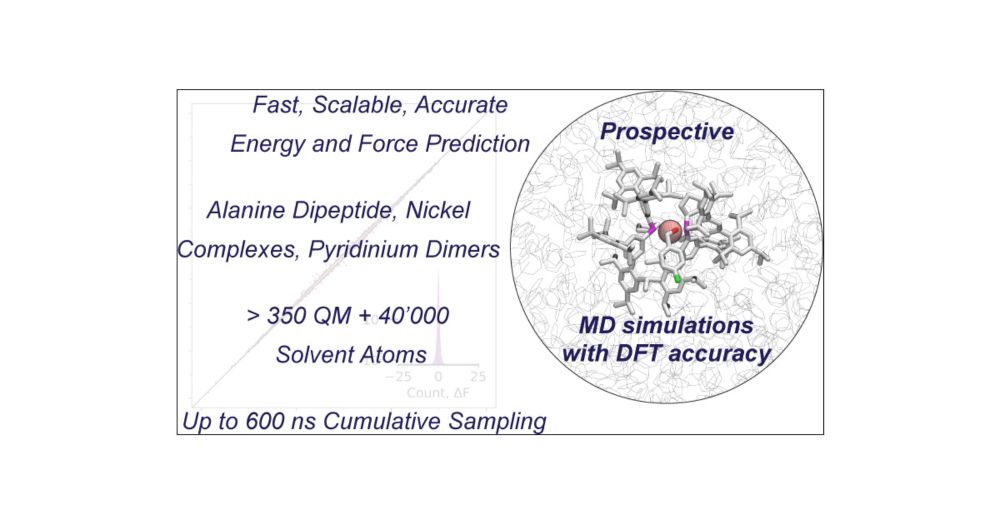
Our new paper in JACS presents the application of a new neural network potential, based on our anisotropic message passing approach, to do QM/MM MD simulations. We achieve chemical accuracy on a few quite different types of chemistry.
doi.org/10.1021/jacs...
#chemsky #compchem #opensource
Hi Marwin. Could you please add us too?
Thanks!
Since it's flexible and can be directly integrated into Python code, we think it's a very useful tool for computational science.
04.07.2024 03:27 — 👍 1 🔁 0 💬 0 📌 0
Our newest preprint describes lwreg, a lightweight system for chemical registration. lwreg has a Python API and makes it easy to store the compound structures you use in your work and the experimental data you generate about them.
chemrxiv.org/engage/chemr...
Our paper introducing an implicit solvation model for organic molecule in water based on a graph neural network has just appeared:
pubs.rsc.org/en/content/a...
#chemsky
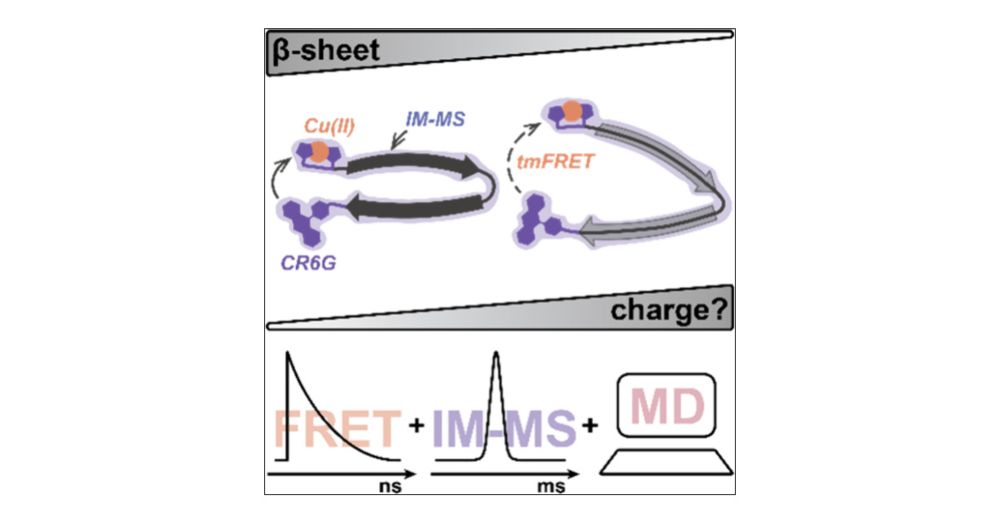
The most recent paper from our ongoing collaboration with the Zenobi group looks at the impact of desolvation on the stability of beta-hairpin structures.
#chemsky
pubs.acs.org/doi/10.1021/...

Our newest preprint, introducing a new type of implicit solvation model based on a graph neural network, is now up: chemrxiv.org/engage/chemr...
09.04.2024 04:18 — 👍 5 🔁 1 💬 0 📌 1
In our newest paper we look at using five different computational methods applied to the results of MD simulations and NMR relaxation experiments in order to better understand protein motions.
doi.org/10.1063/5.01...
Our most recent paper just appeared in JCIM. The title of this one pretty much tells the story: when you assemble a data set by combining data from different literature assays, there is a very good chance that the resulting data contains a lot of noise.
pubs.acs.org/doi/10.1021/...

We have a new preprint out which looks at the amount of noise introduced into a data set when we combine data from different ChEMBL assays.
doi.org/10.26434/che...

Our paper introducing SIMPD is now out. SIMPD is an algorithm for creating training/test sets for molecular #machinelearning based on an analysis of a large number of real-world medchem projects.
link.springer.com/article/10.1...
#opensource code and data are in github.
github.com/rinikerlab/m...

Our most recent preprint describes research done together with the Ferrage group at the Sorbonne in Paris to apply #MolecularDynamics and #NMR to understand protein motions in solution.
chemrxiv.org/engage/chemr...
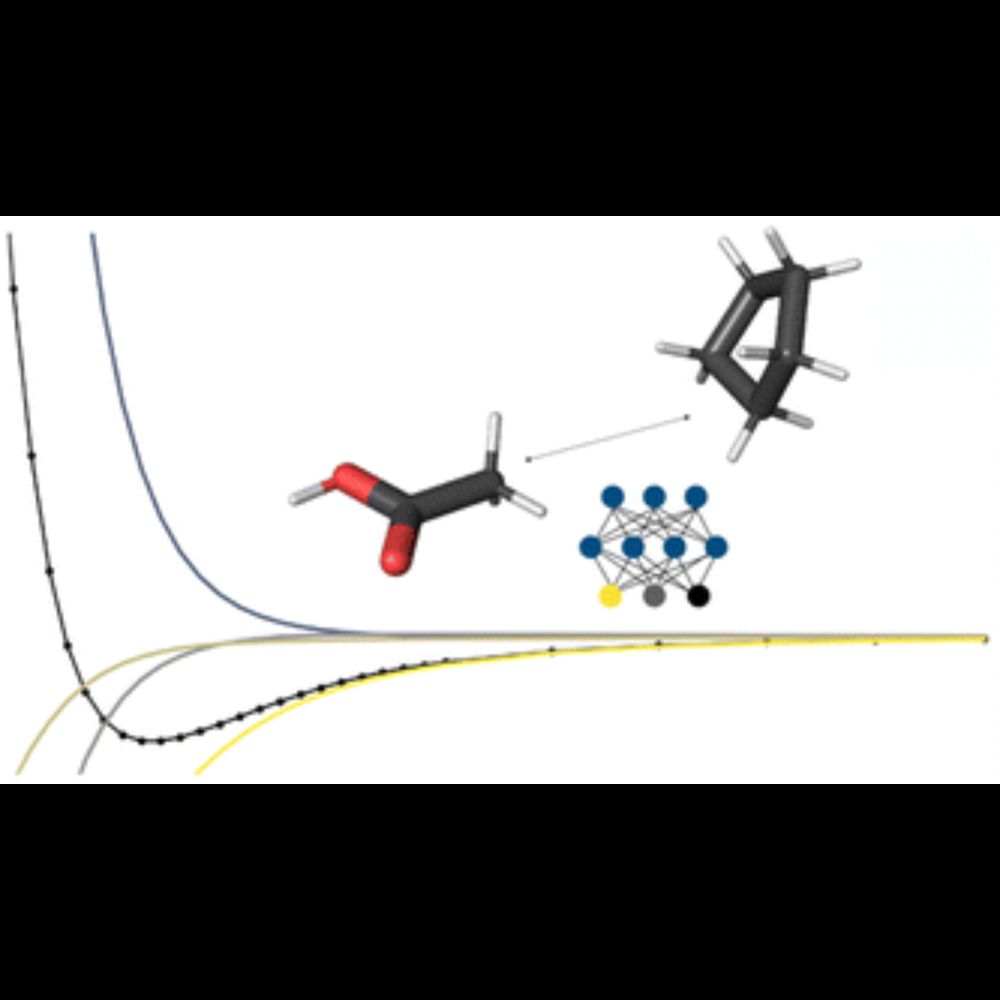
Our most recent publication describes a hybrid classical/machine-learning forcefield we've developed for condensed-phase systems.
As usual, it's #opensource, #opendata, and #openaccess
pubs.rsc.org/en/content/a...
Our paper introducing DASH, an efficient approach for assigning partial charges to atoms in molecules is now out. The method uses a hierarchy created from attention values from a GNN trained on QM data.
It's #opensource, #opendata, and #openaccess
pubs.acs.org/doi/10.1021/...