Genetic regulation of cell type-specific chromatin accessibility shapes immune function and disease risk
Understanding how genetic variation influences gene regulation at the single-cell level is crucial for elucidating the mechanisms underlying complex diseases. However, limited large-scale single-cell multi-omics data have constrained our understanding of the regulatory pathways that link variants to cell type-specific gene expression. Here we present chromatin accessibility profiles from 3.5 million peripheral blood mononuclear cells (PBMCs) across 1,042 donors, generated using single-cell ATAC-seq and multiome (RNA+ATAC) sequencing, with matched whole-genome sequencing, generated as part of the TenK10K program. We characterized 440,996 chromatin peaks across 28 immune cell types and mapped 243,273 chromatin accessibility quantitative trait loci (caQTLs), 60% of which are cell type-specific. Integration with TenK10K scRNA-seq data (5.4 million PBMCs) identified 31,688 candidate cis-regulatory elements colocalized with eQTLs; over half (52.5%) show evidence of causal effects mediated via chromatin accessibility. Integrating caQTLs with GWAS summary statistics for 16 diseases and 44 blood traits uncovered 9.8% - 30.0% more colocalized signals compared with using eQTLs alone, many of which have not been reported in prior studies. We demonstrate cell type-specific mechanisms, such as a regulatory effect on IRGM acting through altered promoter chromatin accessibility in CD8 effector memory T cells but not in naive cells. Using a graph neural network, we inferred peak-to-gene relationships from unpaired multiome data by incorporating caQTL and eQTL signals, achieving up to 80% higher accuracy compared to using paired multiome data without QTL information. This improvement further enhanced gene regulatory network inference, leading to the identification of 128 additional transcription factor (TF)-target gene pairs (a 22% increase). These findings provide an unprecedented single-cell map of chromatin accessibility and genetic variation in human circulating immune cells, establishing a powerful resource for dissecting cell type-specific regulation and advancing our understanding of genetic risk for complex diseases. ### Competing Interest Statement L.C., E.B.D., and K.K.H.F. are employed at Illumina Inc. D.G.M. is a paid advisor to Insitro and GSK, and receives research funding from Google and Microsoft, unrelated to the work described in this manuscript. G.A.F reports grants from National Health and Medical Research Council (Australia), grants from Abbott Diagnostic, Sanofi, Janssen Pharmaceuticals, and NSW Health. G.A.F reports honorarium from CSL, CPC Clinical Research, Sanofi, Boehringer-Ingelheim, Heart Foundation, and Abbott. G.A.F serves as Board Director for the Australian Cardiovascular Alliance (past President), Executive Committee Member for CPC Clinical Research, Founding Director and CMO for Prokardia and Kardiomics, and Executive Committee member for the CAD Frontiers A2D2 Consortium. In addition, G.A.F serves as CMO for the non-profit, CAD Frontiers, with industry partners including, Novartis, Amgen, Siemens Healthineers, ELUCID, Foresite Labs LLC, HeartFlow, Canon, Cleerly, Caristo, Genentech, Artyra, and Bitterroot Bio, Novo Nordisk and Allelica. In addition, G.A.F has the following patents: "Patent Biomarkers and Oxidative Stress" awarded USA May 2017 (US9638699B2) issued to Northern Sydney Local Health District, "Use of P2X7R antagonists in cardiovascular disease" PCT/AU2018/050905 licensed to Prokardia, "Methods for treatment and prevention of vascular disease" PCT/AU2015/000548 issued to The University of Sydney/Northern Sydney Local Health District, "Methods for predicting coronary artery disease" AU202290266 issued to The University of Sydney, and the patent "Novel P2X7 Receptor Antagonists" PCT/AU2022/051400 (23.11.2022), International App No: WO/2023/092175 (01.06.2023), issued to The University of Sydney. ### Funding Statement A.X. is supported by NHMRC Investigator grant 2033018. J.E.P. is supported by NHMRC Investigator grant 2034556, and a Fok Family Fellowship; D.G.M. is supported by an NHMRC investigator grant (2009982). G.A.F. and the BioHEART Study have been supported by NHMRC Investigator Grant, NSW Health Office of Health and Medical Research, and the NSW Health Statewide Biobank scheme. ### Author Declarations I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained. Yes The details of the IRB/oversight body that provided approval or exemption for the research described are given below: The Human Research Ethics Committee of St Vincent's Hospital gave ethical approval for this work. The National Statement on Ethical Conduct in Human Research of the National Health and Medical Research Council gave ethical approval for this work. I confirm that all necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived, and that any patient/participant/sample identifiers included were not known to anyone (e.g., hospital staff, patients or participants themselves) outside the research group so cannot be used to identify individuals. Yes I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance). Yes I have followed all appropriate research reporting guidelines, such as any relevant EQUATOR Network research reporting checklist(s) and other pertinent material, if applicable. Yes Raw caQTL summary statistics will be available at Zenodo website prior to acceptance. [https://github.com/powellgenomicslab/tenk10k\_phase1\_multiome][1] [1]: https://github.com/powellgenomicslab/tenk10k_phase1_multiome
New preprint alert: tinyurl.com/tenk10k-multiome. Excited to share our analysis on the impact of genetic variants on single-cell chromatin accessibility in blood, using scATAC-seq and WGS from over 1,000 donors and 3.5M nuclei as part of TenK10K phase 1 🧬
🧵👇 (1/n)
01.09.2025 11:59 — 👍 17 🔁 12 💬 1 📌 2

NSW CVRN Rising Stars Seminar – 11 Sept
Join us at VCCRI to hear from Dr Albert Henry @alberthenry.bsky.social (Garvan Institute), a rising leader in data science, cardiovascular genetics and systems biology.
📅 Wed 11 Sept | 📍VCCRI
Register now: shorturl.at/r7eWD
18.08.2025 06:33 — 👍 2 🔁 1 💬 0 📌 0
Big thanks to co-first author Anne Senabouth, supervisor Joseph Powell, co-authors Rika Tyebally @blakebowen.bsky.social @lawrencehuang.bsky.social Jayden Fan @petercallen.bsky.social @anglixue.bsky.social @htanudisastro.bsky.social @annasecuomo.bsky.social @dgmacarthur.bsky.social, and many others!
01.09.2025 04:30 — 👍 4 🔁 0 💬 0 📌 0
10. Last but not least, this study would not be possible without massive contributions from all the co-authors and the wider TenK10K team. If you like to learn more about TenK10K, check out our other studies:
tinyurl.com/tenk10k-flagship
tinyurl.com/tenk10k-repeats
tinyurl.com/tenk10k-multiome
01.09.2025 04:30 — 👍 2 🔁 0 💬 1 📌 1
9. We hope our study offers a cell type-resolved map for causal inference of gene expression on complex traits to help understand disease mechanisms and guide drug development.
There are still plenty to unpack. We encourage reading the preprint and would love to hear feedback!
01.09.2025 04:30 — 👍 2 🔁 0 💬 1 📌 0


8. We found 116 genes associated with Crohn’s disease show differential expression in equivalent colon tissue cell types sampled from healthy and diseased individuals in an external dataset. This includes ZBTB38, a candidate susceptibility gene implicated in recent GWAS.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0

7. We highlight an interesting example of cell type-specific causal effect of NCF4 gene in Crohn’s disease, which shows a protective (-) effect in B naive and risk-increasing (+) effect in B memory, implicating context-specific regulation that can be resolved using sc-eQTL MR.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0


6. Drugs targeting gene-trait associations identified through sc-eQTL MR in this study are 3.3 times more likely to have secured regulatory approval. Among these were major targets such as JAK2 & TNF for Crohn’s disease, APP for Alzheimer’s disease, and GLP1R for type 2 diabetes.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0

5. We found different polygenic enrichment patterns amongst dendritic cell (DC) subtypes: Crohn’s disease enrichment were found in cDC1, cDC2, and ASDC subtypes, and COVID-19 found in pDC only - consistent with its function for rapid interferon signalling in viral infection.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0

4. Through single-cell Disease Relevance Score (scDRS) analysis, we found that peripheral immune cells are enriched for polygenic signature of most complex traits, implicating widespread pleiotropy beyond immune function and peripheral blood composition.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0


3. We identified 190,449 gene-trait associations, including 34% not found by gene-level analysis of GWAS data, and 61% not found by MR using whole blood eQTL. Associations found only by sc-eQTL MR are often restricted to fewer cell types, implicating cell type specificity.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0
2. Our study presents a catalogue of cell type-specific causal effects of gene expression on 53 diseases (8,672 genes), and 31 biomarker traits (16,085 genes) across 28 peripheral immune cell types identified using Mendelian randomisation (MR) with sc-eQTL genetic instruments.
01.09.2025 04:30 — 👍 1 🔁 0 💬 1 📌 0
Lastly, it goes without saying that it takes a village to publish this study. I'd like to take this opportunity to thank my previous PhD and postdoc advisor at UCL, Dr. Tom Lumbers who led this project, friends and collaborators within the HERMES Consortium, and all the study participants.
END/
04.03.2025 11:28 — 👍 0 🔁 0 💬 0 📌 0
Genome-wide association study meta-analysis provides insights into the etiology of heart failure and its subtypes
BONUS for those who scroll long enough to find this:
We also have an online supplementary information with more details on:
1. GWAS QC pipeline
2. Locus zoom, gene prioritisation, cross-trait association, and study-level association for each heart failure locus
hermes2-supp-note.netlify.app
14/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0
Datasets | Common Metabolic Diseases Knowledge Portal
We have also released the GWAS summary statistics for browsing and download via the CVD Knowledge Portal:
* Mixed-ancestry meta-analysis:
cvd.hugeamp.org/dinspector.h...
* European ancestry meta-analysis:
cvd.hugeamp.org/dinspector.h...
13/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

We performed genetic correlation (rg) and Mendelian randomisation analyses to distinguish between shared genetics and causal relationships. This is most apparent in CAD and ni-HF, which shows positive rg without causal effect. Interestingly, T2D shows this pattern on all HF subtypes.
11/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

We further explored the extent of pleiotropic effects in HF loci on risk factors and diseases associated with HF. Through colocalisation analysis, we found that HF shared causal genetic variants with at least one of 22 other traits at 42 loci.
10/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

The identified genotype-phenotype clusters provide insights into etiological modules underlying HF pathology, e.g. cluster 1: ischaemic & major cardiovascular disorders, cluster 2: arrythmia & cardiomyopathies, cluster 4: hypertension, cluster 5: metabolic disorders.
9/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

Next, we characterised the downstream effect of lead variants in HF susceptibility loci on 294 human diseases in UK Biobank. We then used network analysis and community detection technique to identify 18 distinct genotype-phenotype clusters from these phenome-wide association results.
8/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

Using sn-RNAseq from 16 healthy & 28 failing heart donors, we found enrichment of cardiomyocyte genes. We also identified 53 GWAS genes that were differentially expressed in cardiac cell types, notably cardiomyocytes and fibroblasts.
7/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

Through heritability enrichment analysis, we found differential involvement of tissues across HF subtypes. Notably, whilst other HF subtypes were mostly enriched for genes that are more specifically expressed in cardiac tissues, ni-HFpEF was distinctly enriched for kidney and pancreas.
6/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

Integrating multiple gene prioritisation strategies, we shortlisted 142 candidate effector genes across 66 genetic loci for HF, and nominated the most likely effector gene for each locus. This includes IGFBP7 for HFpEF, which is linked to cardiomyocyte senescence and cardiac remodelling.
5/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

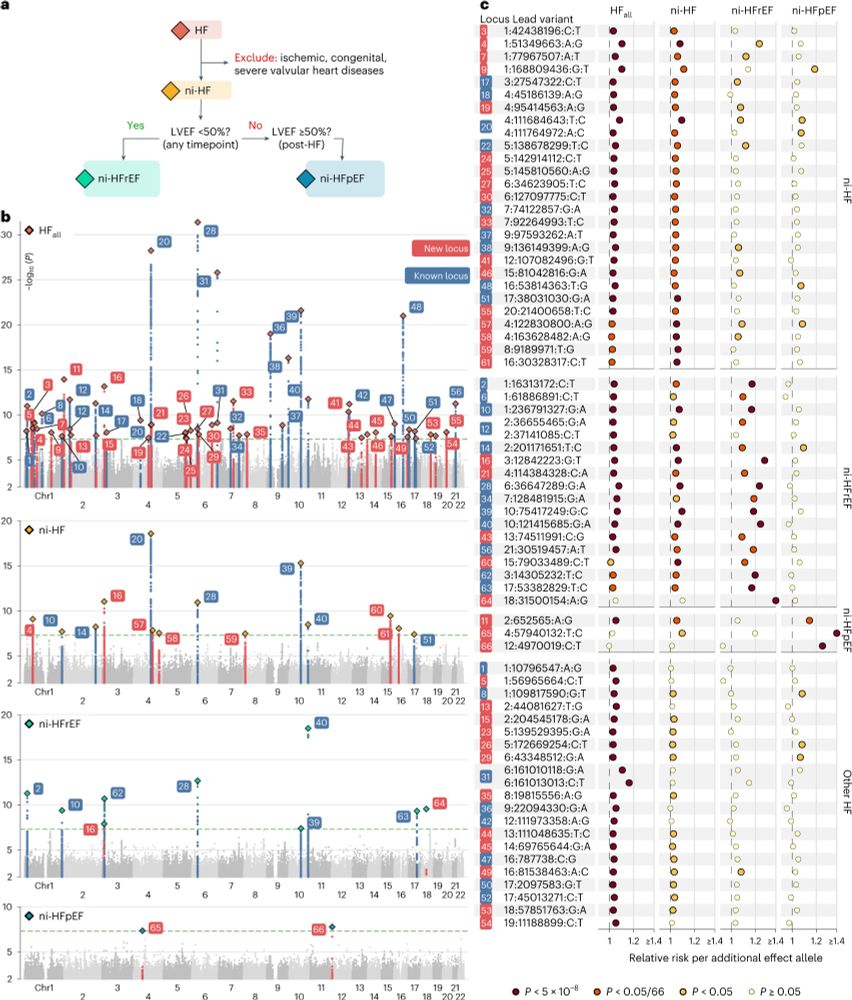
We found 66 independent genetic loci associated with at least 1 HF phenotype, including 37 not previously linked to HF. Of note, 10 / 66 loci were identified in GWAS of ni-HF subtypes despite smaller N compared to HF-all; showing the importance of phenotype definition in a case-control GWAS
4/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0
First, we collected data from 49 GWAS comprising >150k HF cases and performed meta-analyses on 4 subsets of HF phenotypes:
1. Overall HF (HF-all)
2. non-ischaemic HF (ni-HF)
3. ni-HF with reduced (<50%) ejection fraction (ni-HFrEF)
4. ni-HF with preserved (≥50%) ejection fraction (ni-HFpEF)
3/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0

Heart failure (HF) is a complex disease associated with many etiologies and risk factors. Here, we study how genetic variation influence risk of different HF subtypes and integrate these results with other genomic information to uncover disease etiology
2/
04.03.2025 11:28 — 👍 0 🔁 0 💬 1 📌 0
ICYMI:
We delve into the molecular etiology of dilated cardiomyopathy, combining genomics, cellular transcriptomics, and clinical phenotype in population biobanks & clinical cohorts.
Read the full paper below 👇
28.11.2024 06:32 — 👍 2 🔁 0 💬 0 📌 0
🧬 Our dilated #cardiomyopathy GWAS out today!
📍https://www.nature.com/articles/s41588-024-01952-y
All made possible with friends and collaborators from HERMES Consortium @alberthenry.bsky.social @tomlumbers.bsky.social @jamesware.bsky.social @mrc-lms.bsky.social @imperialnhli.bsky.social #BHF
🧵 👇
21.11.2024 13:35 — 👍 43 🔁 23 💬 5 📌 3
Genomics, big data, open science, diversity. Director of the Centre for Population Genomics, focused on building a more equitable future for genomic medicine. Opinions my own.
MD-PhD candidate at the University of Sydney & Garvan Institute.
Studying tandem repeats in single-cell contexts w/ @dgmacarthur.bsky.social
EMBO Postdoctoral fellow at the Garvan Institute of Medical Research, Sydney, Australia.
Previously EMBL-EBI, Wellcome Sanger Institute and University of Cambridge in Cambridge, UK.
All things single-cell, genetics & genomics.
Head of Human Genetics & Senior Group Leader @ Wellcome Sanger Institute. Statistical geneticist working on complex diseases #ibd #firstgen.
Associate Professor @ Big Data Institute, University of Oxford
2024 Lister Institute Fellow
genomics | rare disease | gene regulation | genetic therapies
https://rarediseasegenomics.org/
(field) hockey player | cyclist | hiker
Executive Director EMBL. I have an insatiable love of biology. Consultant to ONT and Cantata (Dovetail)
Co-Founder and Chief Scientist | Venture Partner | Professor | Mentor | Proud mom of 3 ✨
Nature Portfolio’s high-quality products and services across the life, physical, chemical and applied sciences is dedicated to serving the scientific community.
NYT columnist. Signal: carlzimmer.51
Newsletter: https://buttondown.com/carlzimmer/
Web: http://carlzimmer.com
[This account includes a tweet archive]
Behavior genetics, clinical psychology, Mets. Occasional politics.
Substack (free): https://ericturkheimer.substack.com/
Book Is Out! Understanding the Nature-Nurture Debate
https://shorturl.at/Ce2hf
Electronic Version: https://shorturl.at/Fq2jv
Assistant professor in quantitative and computational biology @USC. Genetics, evolution, statistics. https://edgepopgen.github.io/edgelab/
Population and evolutionary genetics @UCDavis. Posts, grammar, & spelling are my views only. He/him. #OA popgen book https://github.com/cooplab/popgen-notes/releases
Statistical geneticist. Associate Prof at Dana-Farber / Harvard Medical School.
www.gusevlab.org
My lab at Stanford studies human population genetics and complex traits.
Epidemiologist + Statistician | Clinical Research Facility - University College Cork | UCC School of Public Health | #ClinicalTrials #Epidemiology #Statistics #RStats #WBE #IDSurveillance
Views mine -> https://statsepi.substack.com/
statistician • associate prof • team lead health data science and head methods research program at julius center • director ai methods lab, umc utrecht, netherlands • views and opinions my own
Professor of Biostatistics
Vanderbilt University School of Medicine
Expert Biostatistics Advisor
FDA Center for Drug Evaluation and Research
https://hbiostat.org https://fharrell.com
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app