I think Bluesky is a great platform and I’m here to stay. However, a bookmark function would make the experience SO much better and much more functional for research/ information gathering, especially for reporters. Pretty please @support.bsky.team @bsky.app ?
14.01.2025 20:14 — 👍 11780 🔁 1112 💬 402 📌 89

Fig. 1: The Chromosome number and Hidden State-dependent Speciation and Extinction (ChromoHiSSE) model. Panel (a) describes the event rates allowed in the model for both cladogenetic (upper) and anagenetic (lower) events. Panels (b) and (c) demonstrate those rates on a tree simulated under ChromoHiSSE. Branches that do not reach the present represent extinct, unsampled lineages. Panel (b) shows the anagenetic and cladogenetic changes in the hidden states, indicated by blue (i) vs orange (ii). Panel (c) shows the anagenetic and cladogenetic gains and losses of chromosomes, as well as speciation with no corresponding change in chromosome numbers. More red colors in (c) correspond to more chromosomes. The vertical blue and orange bars in (c) indicate clades in the blue (i) vs orange (ii) hidden states, displaying that chromosome number changes are less frequent in the blue hidden state than in the orange. The asterisk in (c) demarcates where a cladogenetic dysploidy event could appear as an anagenetic event because of unsampled lineages.
New study by @tribblelab.bsky.social @jimarcor.bsky.social @marcialescudero.bsky.social Michael May, Rosana Zenil-Ferguson and myself just out:
Introduces a novel HiSSE chromosome model and demonstrates the importance of chrom. evol. in #sedge diversity.
nph.onlinelibrary.wiley.com/doi/10.1111/...
26.12.2024 13:01 — 👍 75 🔁 24 💬 1 📌 1

Instead of listing my publications, as the year draws to an end, I want to shine the spotlight on the commonplace assumption that productivity must always increase. Good research is disruptive and thinking time is central to high quality scholarship and necessary for disruptive research.
20.12.2024 11:18 — 👍 1155 🔁 375 💬 21 📌 57
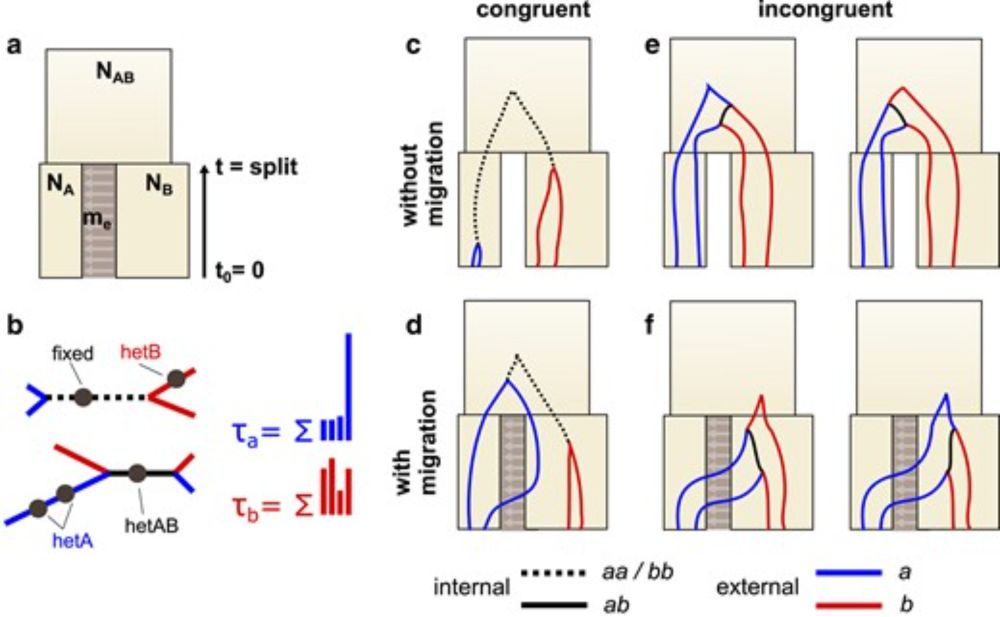
Genealogical asymmetry under the isolation with migration model and a two-taxon test for gene flow
Abstract. Methods for detecting gene flow between populations often rely on asymmetry in the average length of particular genealogical branches, with the A
My and Derek Setter's work on gene flow is now published in the latest issue of Genetics. We show that it is possible to detect past gene flow between two populations by summarising the asymmetry in pop-specific external branch lengths. 1/2
tinyurl.com/5ff8ch9e
12.12.2024 14:17 — 👍 62 🔁 34 💬 1 📌 0
Climate drives long-term landscape and rapid short-term promoter evolution in the Western Canaries lizard, Gallotia galloti https://www.biorxiv.org/content/10.1101/2024.12.18.628907v1
21.12.2024 11:32 — 👍 0 🔁 1 💬 1 📌 0
Structural rearrangements and selection promote phenotypic evolution in Anolis lizards https://www.biorxiv.org/content/10.1101/2024.12.12.628123v1
17.12.2024 05:33 — 👍 5 🔁 3 💬 1 📌 0
macOS folk: This is your periodic reminder 🎗️ to:
```bash
$ brew update && brew upgrade && brew cleanup
```
I just got back 10.7 GB & patched a slew of CVEs.
16.12.2024 01:55 — 👍 875 🔁 65 💬 42 📌 7

New paper with @snaildit.bsky.social❗
You can use paralogs to reduce long-branch attraction!
This will be especially helpful when there are no extant taxa that can be sampled to break up these branches.
www.biorxiv.org/content/10.1...
13.12.2024 14:35 — 👍 84 🔁 35 💬 2 📌 3
I would also like to recommend @melisaolave.bsky.social
08.12.2024 15:23 — 👍 1 🔁 0 💬 0 📌 0
Hi Joana, I would be happy to be included in the list!
08.12.2024 15:22 — 👍 1 🔁 0 💬 1 📌 0
Getting started with elmer
If you're interested in trying out LLMs in #rstats but don't know where to begin, I've added a few two vignettes to elmer: elmer.tidyverse.org/articles/elm... and elmer.tidyverse.org/articles/pro...
29.11.2024 15:45 — 👍 265 🔁 64 💬 10 📌 1

Gene behind orange fur in cats found at last
After 60 years, scientists know why gingers, calicos, and tortoiseshells look the way they do
Sara Reardon's story on the genetics of coat color in cats was a delight to read (and not just because my sweet calico, Mona Lisa, got to make her Science debut). Now I'm just waiting for scientists to make a discovery about tabbies, so Mo's brother Vinny can get his moment in the spotlight!
27.11.2024 21:17 — 👍 17 🔁 6 💬 2 📌 0

NEW Publication!😄
Here, we show how #PhyKIT — a broadly applicable toolkit for #phylogenomics — can be used to construct phylogenomic data matrices (& quantify biases therein), detect anomalies in predicted orthology, calculate gene-gene coevolution, & more!
🔗: tinyurl.com/yc5ja4xz
25.11.2024 17:25 — 👍 164 🔁 63 💬 2 📌 2
Migrando a 🦋
24.11.2024 03:22 — 👍 1 🔁 0 💬 0 📌 0
Bluesky account of the Open Consortium of Squamate Genomics (OCSG) 🦎🐍
Bióloga, casi investigadora del CONICET. Hormigas (🐜) is my passion. 🇦🇷 postdoc in 🇩🇪.
“¿Quién se pregunta todavía por el valor de una ciencia que consume a sus criaturas de una forma tan vampírica?” F. Nietzsche
Argentinean ecologist at IPEEC CONICET 🇦🇷 and fungi lover 💜🍄🟫, mostly interested in microbial ecology 🦠 and plant-soil interactions 🍄🌲. Also passionate about cooking 👨🏼🍳, acting 🎭 and traveling 🌎
AE 📑: J. Appl. Ecol / Biol. Invasions
Investigador Principal CONICET
Grupo de Ecología en Ambientes Costeros - IPEEC
Editor Asociado para Biological Invasions (First Records Collection)
Editor Asociado para Aquatic Invasions
Consejo Asesor Editorial para Ambio
GBE publishes leading original research at the interface between evolutionary biology and genomics.
🔗 academic.oup.com/gbe
🏠 @official-smbe.bsky.social
🤝 @molbioevol.bsky.social
#genome #evolution #science #biology #societyjournal
Into evolution, genetics, reproduction. Immigrant (actual, and from the bird site). She/her.
Ramen addicted evolutionary biologist working on genomics, ecology, speciation and adaptation in Passer sparrows. Associate Professor based in CEES/EVOGENE at the University of Oslo
Biologist | Eco-Evo and sometimes Dynamics | @ILIM • Res. Dept. for Limnology, Mondsee • Univ. of Innsbruck
Evolutionary Biologist | Genomics of Speciation + Adaptation 🧬🦅🐟 | Curator Vertebrates @Natural History Museum Basel & PI @Uni Basel🇨🇭| Views my own | He/him.
Theoretician, Speciation, Sexual selection, will post rarely!
Genetics & Evolution, Faculty at Stanford & Freeman Hrabowski Scholar at HHMI
schumerlab.com
natural historian, PhD Texas, native Londoner. https://www.ucl.ac.uk/taxome/jim (Profile pic from Mauro Cutrona https://facebook.com/profile.php?id=100006219407348)
Evolutionary biology, reproduction, speciation and the odd photo
Genomics, insects and evolution. Particularly butterflies, crop pests and black soldier flies. University of Cambridge, St John’s College and Dept Zoology
PhD student in evolutionary genomics at Uppsala University. Primates, speciation, hybridization, etc. Birder.
Postdoc at @ ETH Zürich; evolution and speciation in flowers (previously fishes); she/her; 🏳️🌈
Associate Prof. @unibern.bsky.social in Theoretical Ecology and Evolution | she/her | Views are my own







