Was amazing to work with all the participants. Had a great week and I hope people learned something too :)
Thanks again to all participants and @physaliacourses.bsky.social for organizing .
Was amazing to work with all the participants. Had a great week and I hope people learned something too :)
Thanks again to all participants and @physaliacourses.bsky.social for organizing .


Today we kicked off the #ComparativeGenomics course with @sedlazeck.bsky.social & Ingo Ebersberger!
On Day 1, we’re covering de novo assembly strategies, best practices, and quality control for genome assemblies.
Sensational #ASHG event on detecting variants, including Structural Variants, from long reads learning.ashg.org/products/unc...
@sedlazeck.bsky.social @bcmhouston.bsky.social
Last seats available! 🚨
Join our #ComparativeGenomics course with @sedlazeck.bsky.social & @Ingo Ebersberger and learn how to analyse, compare, and interpret genomes using modern bioinformatics approaches. Don’t miss it—registration is closing soon!
@nanoporetech.com @pacbio.bsky.social

Rayan and I had some nice dinner too :)
10.02.2026 18:50 — 👍 3 🔁 0 💬 1 📌 0I clearly have to come back and give another talk ... Hahaha
07.02.2026 18:35 — 👍 3 🔁 0 💬 0 📌 0Scalable and comprehensive mosaic variant calling using DRAGEN https://www.medrxiv.org/content/10.64898/2026.02.03.26345450v1
04.02.2026 23:40 — 👍 1 🔁 1 💬 0 📌 0Thanks:)
03.02.2026 07:53 — 👍 2 🔁 0 💬 0 📌 0
We’re excited to confirm Fritz Sedlazeck as our first speaker for #nanoporeconf! His research confronts genomic inequity, identifying novel variants that could influence disease risk, gene regulation and healthcare in Hispanic populations. https://bit.ly/4sOxMKd
21.01.2026 18:55 — 👍 9 🔁 3 💬 0 📌 0
The next edition of our #ComparativeGenomics course is just around the corner (📅 23–27 February)!
A few seats are still available — don’t miss the opportunity to learn comparative #genomics from Ingo Ebersberger & @sedlazeck.bsky.social 🚀
🔗 www.physalia-courses.org/courses-work...


Many thanks to @iscb.bsky.social for a fantastic visit at their meeting in Hong Kong last week! Met so many great people during that week.
Many thanks also to Ruibang ! Always great to make a bad joke while taking a picture ;)
Thanks Ben for a fantastic visit! Hopefully see you soon
05.12.2025 02:05 — 👍 1 🔁 0 💬 0 📌 0

Huge thanks to @sedlazeck.bsky.social l Luis and everyone who joined our Structural Variant Detection & Comparison course! 🚀✨
We dived deep, learned tons, and had some insightful questions and lively discussions — you all made it very successful!
Wishing you all the best smashing your projects!

@benlangmead.bsky.social giving a talk about rowing his boat and explaining pangenomes with that :).
Amazing talk about reference biases and the future to avoid it.
Fascinating talk at @bcmhgsc.bsky.social @riceuniversity.bsky.social ! @treangen.bsky.social




Check major accomplishments of @gregor-research.bsky.social. @eurekalert.bsky.social @bcmhgsc.bsky.social @moezdawood.bsky.social #LupskiLab @sedlazeck.bsky.social @poseypod.bsky.social @bcmhouston.bsky.social S. Montgomery @stanfordmedicine.bsky.social @nature.com www.eurekalert.org/news-release...
20.11.2025 20:56 — 👍 2 🔁 3 💬 0 📌 0

We have just started the workshop on Structural Variant detection from short @illumina and long reads @nanoporetech.com @pacbio.bsky.social with @sedlazeck.bsky.social , Luis and a very international group of attendees.
www.physalia-courses.org/courses-work...

Excited to co-host @benlangmead.bsky.social at @bcmhouston.bsky.social Dec 3rd 4:00-5:00 PM. He is a lead in pangenomics! A rare opportunity to learn more about reference issues, so dont miss out!
More information events.rice.edu/event/guest-...
@treangen.bsky.social @ricecompsci.bsky.social

Join us on our 3 day (Dec 1-3) #workshop @physaliacourses.bsky.social teaching about Structural Variant detection from short and long reads. We will give insights into different approaches to detect SV from somatic to population scale. Only a few places left: www.physalia-courses.org/courses-work...
20.11.2025 11:33 — 👍 5 🔁 1 💬 0 📌 0
Learn about the major accomplishments of @gregor-research.bsky.social. R. Gibbs, @bcmhgsc.bsky.social @moezdawood.bsky.social #LupskiLab @sedlazeck.bsky.social @poseypod.bsky.social @bcmhouston.bsky.social S. Montgomery @stanfordmedicine.bsky.social @nature.com blogs.bcm.edu/2025/11/18/f...
18.11.2025 21:04 — 👍 2 🔁 4 💬 0 📌 0
From coffee-fueled coding marathons ☕ to new genomics tools: Our 2024 #Hackathon paper is out in @f1000publishing.bsky.social !
Huge thanks to everyone world wide who joined the @bcmhgsc.bsky.social madness 🎉
🔗 f1000research.com/articles/14-...
@gregor-research.bsky.social @smahtnetwrk.bsky.social

New in @nature.com! “GREGoR: Accelerating Genomics for Rare Diseases” highlights how the GREGoR Consortium is advancing rare disease discovery through data sharing, multi-omics, and next-gen sequencing across 7,500+ individuals in 3,000+ families.
🧬 www.nature.com/articles/s41...

From coffee-fueled coding marathons ☕ to new genomics tools: Our 2024 #Hackathon paper is out in @f1000publishing.bsky.social !
Huge thanks to everyone world wide who joined the @bcmhgsc.bsky.social madness 🎉
🔗 f1000research.com/articles/14-...
@gregor-research.bsky.social @smahtnetwrk.bsky.social
This is a great course and deep dive into comp. genomics with Ingo and me! Always great discussions across all types of organisms and how to identify variants and compare assemblies etc. Join us to learn more!
10.11.2025 14:13 — 👍 1 🔁 0 💬 0 📌 0
plsRT: Looking for a motivated postdoc!
Join us at @bcmhgsc.bsky.social to explore the mosaic & somatic landscape of the human genome: structural variants, methylation, and all things @smahtnetwrk.bsky.social
If you like long reads, complex variants & methylation come talk to me!

Join our online #ComparativeGenomics course with Ingo & @sedlazeck.bsky.social , 23-27 Feb 2026! Learn genome assembly, variant detection (SNVs & SVs), and functional impact analysis with hands-on sessions.
www.physalia-courses.org/courses-work...
#Bioinformatics #Genomics

Trio-barcoded @nanoporetech.com Adaptive Sampling (TBAS) to improve #RareDisease diagnostic at less than 1/2 the $$ & high cov: 76% solve rate across 13 trios inc. two corrections from prev. diagnosis. www.medrxiv.org/content/10.1...
@gregor-research.bsky.social @bcmhgsc.bsky.social
#Research

We introduce trio-barcoded
@nanoporetech.com
adaptive sampling (TBAS), which allows targeted sequencing of one trio in one flowcell. We tested it on HG002-4+13 trios from GREGoR.
TBAS and pipeline accurately report causative SNV, SV&TRs. @sedlazeck.bsky.social
www.medrxiv.org/content/10.1...
Working with @sedlazeck.bsky.social and his group on exploring the utility of Constellation has been amazing. So happy to see this preprint out!
21.10.2025 13:12 — 👍 2 🔁 1 💬 1 📌 0Thanks Mitch for all your support!
22.10.2025 16:57 — 👍 1 🔁 0 💬 0 📌 0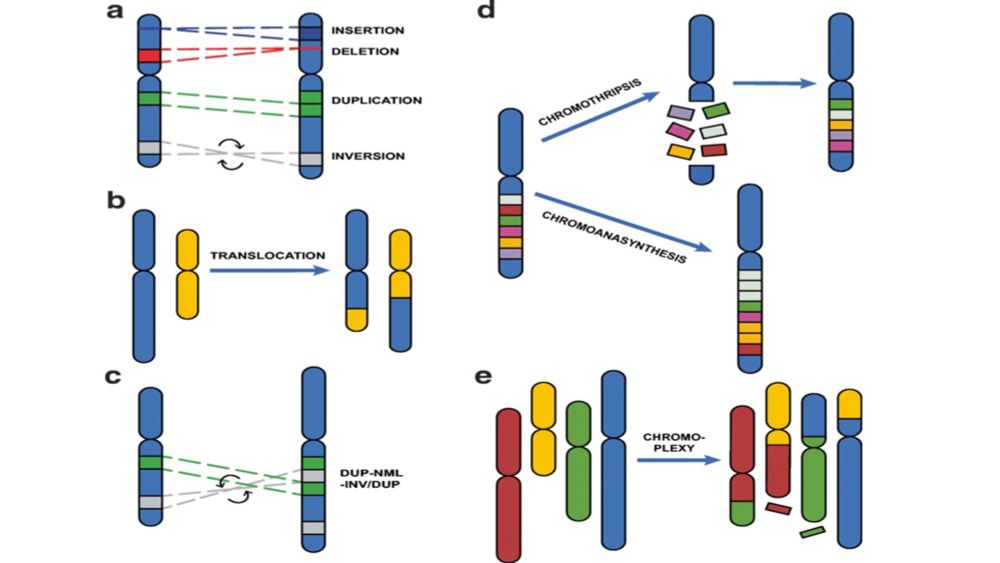
Are you interested in Structural Variants? 🧬 Join our online course (1–3 Dec) to learn how to detect, compare, and annotate SVs from short & long @nanoporetech.com reads with @sedlazeck.bsky.social and Luis Paulin! 🚀
www.physalia-courses.org/courses-work...