
Isobaric labeling with the @thermofishersci.bsky.social TMT kit is super useful in proteomics! 🧬🔥
#Proteomics #TMT #MassSpec #ScienceLife
@map-proteomique.bsky.social
MaP offers since 2007 a wide panel of techniques for protein identification and quantification for basic and clinical research but also for industry.

Isobaric labeling with the @thermofishersci.bsky.social TMT kit is super useful in proteomics! 🧬🔥
#Proteomics #TMT #MassSpec #ScienceLife

Isobaric labeling with the @thermofishersci.bsky.social TMT kit is super useful in proteomics! 🧬🔥
#Proteomics #TMT #MassSpec #ScienceLife
I will be at the 𝐃𝐈𝐀-𝐍𝐍 session @ 𝐓𝐡𝐞𝐫𝐦𝐨 𝐏𝐫𝐨𝐭𝐞𝐨𝐦𝐞 𝐃𝐢𝐬𝐜𝐨𝐯𝐞𝐫𝐞𝐫 & 𝐂𝐨𝐦𝐩𝐨𝐮𝐧𝐝 𝐃𝐢𝐬𝐜𝐨𝐯𝐞𝐫𝐞𝐫 User Meetings in Bremen, Germany, 𝟗-𝟏𝟏 𝐃𝐞𝐜𝐞𝐦𝐛𝐞𝐫. With some cool advances for DIA-NN on Thermo instruments. www.thermofisher.com/de/de/home/e...
01.10.2025 07:19 — 👍 9 🔁 2 💬 0 📌 0
Great to welcome Sébastien JeanJean, Senior Sales Manager France at @thermofishersci.bsky.social, to visit our new #OrbitrapAstral mass spectrometer installation today!
Big thanks to our funders — CPER, LNCC, @ibisa.net, Amidex, and @inserm.fr.
#MassSpec #Proteomics @insermpacacorse.bsky.social

Great to welcome Sébastien JeanJean, Senior Sales Manager France at @thermofishersci.bsky.social, to visit our new #OrbitrapAstral mass spectrometer installation today!
Big thanks to our funders — CPER, LNCC, @ibisa.net, Amidex, and @inserm.fr.
#MassSpec #Proteomics @insermpacacorse.bsky.social
In DIA-NN 2.3.0, we have added Deamidation (NQ) as an option in the GUI. There are good reasons for it :) You can try InfinDIA with any number of modifications selected, and check the RT differences between modified vs respective stripped peptides, for each mod - a way to validate peptidoform FDR.
29.09.2025 11:46 — 👍 13 🔁 3 💬 0 📌 0
With 𝗗𝗜𝗔-𝗡𝗡 𝟮.𝟯.𝟬 Preview (Academia-only for now), we showcase the transformative new capabilities that have been developed in the past months. Download: github.com/vdemichev/Di...
26.09.2025 09:52 — 👍 29 🔁 6 💬 3 📌 0
🚨Publication alert🌶️: #Immunopeptidomics is a trending for good reason! A team from @ki.se identify ovarian tumor-derived peptide that induces memory-like features in natural killer cells, paving the way for new cancer immunotherapy strategies! www.biorxiv.org/content/10.1...
23.09.2025 12:35 — 👍 3 🔁 1 💬 0 📌 0
Are you using DIA mass spectrometry in proteomics?
We have just put this paper online: www.biorxiv.org/content/10.1...
Here, we came up with some Recommendations for Quantitative Data-Independent Acquisition (DIA) Proteomics using Controlled Quantitative Experiments (CQEs)
1/4
Proteomics Webinar: DIA with FragPipe, DIA-NN, and Skyline
Presenters: Eduard Sabidó and Brendan MacLean
When: Tuesday, September 16, 8am (Pacific Time)
Register Now ... skyline.ms/project/home...
#massspec #proteomics

@kentsisresearch.bsky.social develop a proteogenomics workflow to detect 1000s of unknown cancer proteins. PG3 combines long-read RNA sequencing with multi-protease MS to discover non-canonical proteins in cancer cells. Used a timsTOF HT, nanoElute 2 & Aurora Ultimate 25x75 column:
bit.ly/4mlQSmu

The key molecules in immune responses are glycoproteins.
Why isn't glycosylation studied more intensively?
It should be.
It's a challenging research area, but a good one for ambitious & audacious scientists.
Time and time again, it's always the same picture. DIA-NN controls FDR correctly as data reliability has been the main goal at DIA-NN's conception back in 2017 and a priority since then.
25.07.2025 11:22 — 👍 14 🔁 5 💬 1 📌 0In fact, two essential companions were installed: the Evosep ENO @evosep.bsky.social and the Ionbench table #IonBench
23.07.2025 16:37 — 👍 3 🔁 2 💬 0 📌 0
A new companion for our mass spectrometer.
23.07.2025 06:46 — 👍 1 🔁 0 💬 0 📌 1
Today is a great day!
Our article accepted on March 19, 2025 is published in Wiley mLife journal.
Great collaborative work with JM Bolla's lab, @map-proteomique.bsky.social @canaanlab.bsky.social
Céline Crauste from @ibmm-balard.bsky.social, and more...
👉 onlinelibrary.wiley.com/doi/10.1002/...

At the European Proteomics Congress in St Malo #EuPA2025, Marseille Proteomique @map-proteomique.bsky.social won 3rd prize in the sandcastle competition sponsored by @brukercorporation.bsky.social
20.06.2025 06:05 — 👍 8 🔁 2 💬 0 📌 0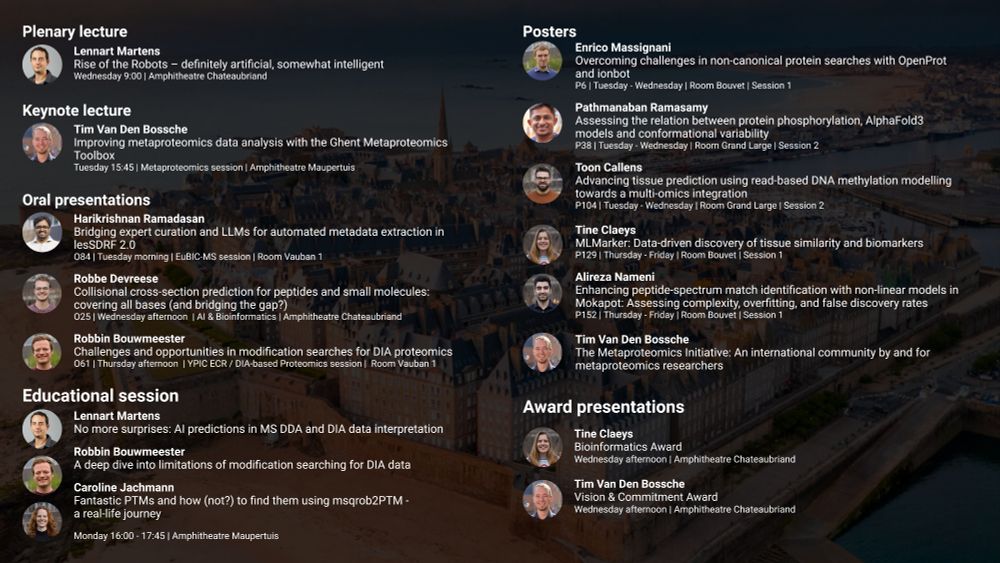
Plenary talk Lennart Martens Rise of the Robots – definitely artificial, somewhat intelligent Keynote lecture Tim Van Den Bossche Improving metaproteomics data analysis with the Ghent Metaproteomics Toolbox Oral presentations Harikrishnan Ramadasan Bridging expert curation and LLMs for automated metadata extraction in lesSDRF 2.0 Robbe Devreese Collisional cross-section prediction for peptides and small molecules: covering all bases (and bridging the gap?) Robbin Bouwmeester Challenges and opportunities in modification searches for DIA proteomics Educational session Lennart Martens No more surprises: AI predictions in MS DDA and DIA data interpretation Robbin Bouwmeester A deep dive into limitations of modification searching for DIA data Caroline Jachmann Fantastic PTMs and how (not?) to find them using msqrob2PTM - a real-life journey Poster presentations Enrico Massignani Overcoming challenges in non-canonical protein searches with OpenProt and ionbot Pathmanaban Ramasamy Assessing the relation between protein phosphorylation, AlphaFold3 models and conformational variability Toon Callens Advancing tissue prediction using read-based DNA methylation modelling towards a multi-omics integration Tine Claeys MLMarker: Data-driven discovery of tissue similarity and biomarkers Alireza Nameni Enhancing peptide-spectrum match identification with non-linear models in Mokapot: Assessing complexity, overfitting, and false discovery rates Tim Van Den Bossche The Metaproteomics Initiative: An international community by and for metaproteomics researchers Award presentations Tine Claeys Bioinformatics Award Tim Van Den Bossche Vision & Commitment Award
From PTMs to proteins, from metadata to metaproteomics. CompOmics has got you covered at #EuPA2025!
16.06.2025 08:05 — 👍 22 🔁 8 💬 0 📌 0
On the way to the European #proteomics association #EuPA2025 conference in St Malo
16.06.2025 09:10 — 👍 3 🔁 1 💬 0 📌 0Our new mass spectrometer has come to life. Now we're going to have to feed it protein several times a day.
03.06.2025 06:47 — 👍 4 🔁 1 💬 0 📌 0
Marseille Proteomics @map-proteomique.bsky.social begins is delighted to announce that our new spectrometer is now out of the box and will start running next week
23.05.2025 14:46 — 👍 2 🔁 2 💬 0 📌 0
A new era for Marseille Proteomics have just reached 100 subscribers to @map-proteomique.bsky.social begins with the arrival of these boxes containing new generations of equipment.
23.05.2025 09:16 — 👍 2 🔁 2 💬 0 📌 0
A new era for Marseille Proteomics have just reached 100 subscribers to @map-proteomique.bsky.social begins with the arrival of these boxes containing new generations of equipment.
23.05.2025 09:16 — 👍 2 🔁 2 💬 0 📌 0
🚨 New preprint! 🚨
Presenting diaPASEF immunopeptidomics for bacterial epitope discovery.
💥 Showcasing DIA-NN immunopeptidomics using proteome-wide predicted HLA class I libraries!
🧬 Read more: biorxiv.org/cgi/content/...
#Immunopeptidomics #MassSpec #DIA #HLA

May the 4th be with you! Celebrate Star Wars Day with the force of Evotip Pure, the trusted ally for your LC-MS journeys!
#MayThe4th #StarWarsDay #LCMS

Exciting news: We have released #FragPipe 23, and it's one of our biggest updates ever. Windows installer, support for TMT on Astral and timsTOF, TMT35, PTM site reports for DIA, improved Astral data handling in #MSFragger, improved diaTracer for diaPASEF data, better Skyline integration, and more!
05.05.2025 04:06 — 👍 77 🔁 15 💬 4 📌 1
Marseille proteomics joins #starwars Day to explore the universe of the proteome on a new dimension. May the fourth be with you
03.05.2025 22:03 — 👍 4 🔁 2 💬 0 📌 0
Marseille proteomics joins #starwars Day to explore the universe of the proteome on a new dimension. May the fourth be with you
03.05.2025 22:03 — 👍 4 🔁 2 💬 0 📌 0
Thank you to all our subscribers. We have just reached 100 subscribers to @map-proteomique.bsky.social
29.04.2025 17:05 — 👍 2 🔁 2 💬 0 📌 0We're always delighted to work together on these exciting projects @jefcavalier.bsky.social @canaanlab.bsky.social
29.04.2025 16:59 — 👍 3 🔁 3 💬 0 📌 0