
Supervisors and Projects
Please re-post: If you know (or are!) somebody who might fancy doing a PhD (Oct 2026 start) in my group @oxfordbiochemistry.bsky.social, working on chromatin evolution in prokaryotes (or other things we're interested in), please have a look at www.bioch.ox.ac.uk/supervisors-...
01.09.2025 09:27 — 👍 42 🔁 50 💬 0 📌 1

What an incredible end to the Eukaryotic RNA Turnover meeting in Brighton! Fantastic talks, inspiring speakers, and amazing to meet so many new RNA enthusiasts. Thanks @biochemsoc.bsky.social for a great conference! #BiochemEvent
29.08.2025 13:41 — 👍 7 🔁 2 💬 0 📌 0

Fantastic day at the Eukaryotic RNA Turnover meeting!
We wrapped up insightful sessions on:
- RNA therapeutics & RNA modification
- RNA molecular machines & RNA-binding proteins
- RNA localization & turnover
Thanks to all speakers! @biochemsoc.bsky.social #BiochemEvent
28.08.2025 15:16 — 👍 5 🔁 1 💬 0 📌 0

What a beautiful morning by the Brighton sea to kick off another exciting session at the Eukaryotic RNA Turnover meeting! ☀️🌊 @biochemsoc.bsky.social #BiochemEvent
28.08.2025 08:47 — 👍 3 🔁 1 💬 0 📌 0
Looking forward to presenting my latest work at the Eukaryotic RNA Turnover meeting in Brighton, organised by the Biochemical Society! Excited to connect with fellow researchers. @biochemsoc.bsky.social #BiochemEvent
www.eventsforce.net/biochemsoc/f...
26.08.2025 21:00 — 👍 4 🔁 1 💬 0 📌 0
🚨 Last day to submit your abstracts!
Don’t miss your chance to present at the Londonomics Symposium 2025 🧬✨
18.08.2025 15:54 — 👍 0 🔁 0 💬 0 📌 0
📣 ABSTRACT DEADLINE EXTENDED UNTIL 18TH AUGUST!
Register here: register.oxfordabstracts.com/event/75604?...
Abstract submission: app.oxfordabstracts.com/stages/79029...
15.08.2025 13:22 — 👍 1 🔁 1 💬 0 📌 1
📣 ABSTRACT DEADLINE EXTENDED UNTIL 18TH AUGUST!
15.08.2025 12:56 — 👍 1 🔁 0 💬 0 📌 0
Great work by @romainstrock.bsky.social and the @tobiaswarnecke.bsky.social 's lab showing that some archaea make enzymes that attack bacteria, suggesting archaeal–bacterial conflicts are common.
15.08.2025 12:55 — 👍 1 🔁 0 💬 0 📌 0
congrats!!! 🥳 so nice to see the paper out!
15.08.2025 12:41 — 👍 1 🔁 0 💬 0 📌 0

Are you an Early Career Researcher in bioinformatics? Then this symposium is for you 💡
Join us for a day of talks, networking and career discussions. Present your work to get fresh new ideas and the chance to win prizes 💸
Featuring @avsecz.bsky.social of Google DeepMind as our keynote speaker⚡️
29.07.2025 11:15 — 👍 4 🔁 4 💬 1 📌 7

Huge thanks to everyone who made the RNA conference in San Diego such a great experience! Grateful for all the insightful conversations—especially the thoughtful suggestions during the poster session. Feeling inspired! #RNA2025 #RNA25
03.06.2025 19:37 — 👍 9 🔁 1 💬 0 📌 0
Excited to be heading to San Diego for the annual RNA conference! Looking forward to reconnecting with familiar faces. Hope to see you there!
23.05.2025 14:18 — 👍 2 🔁 0 💬 0 📌 0
Londonomics has made the move over to Bluesky 🦋
We now have 200+ members and we’re excited to bring you together at workshops, networking events, and the annual symposium! 💡
We’ll be kicking things off with a NextFlow workshop in March 👀
11.02.2025 15:35 — 👍 3 🔁 3 💬 0 📌 0
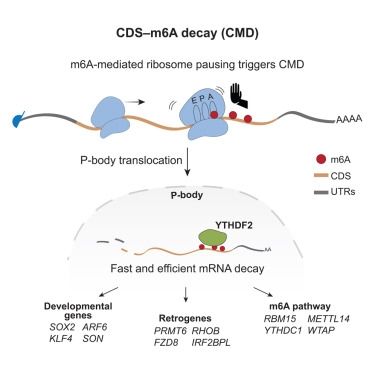
Check out our recent work on CDS #m6A mediated RNA decay (CMD)! Decay relies on ribosome pausing at m6A containing codons. Great collab with the Zarnack lab & colleagues. Thank you @uniwuerzburg.bsky.social , @imbmainz.bsky.social and RMap🥳
www.cell.com/molecular-ce...
15.01.2025 10:00 — 👍 48 🔁 18 💬 0 📌 0
Great work by @sviatoslavsidorov.bsky.social !
02.12.2024 15:48 — 👍 5 🔁 3 💬 0 📌 0
Hi could you add me to lncRNA list. cheers
25.11.2024 09:07 — 👍 2 🔁 0 💬 1 📌 0
could you add me please. cheers!
25.11.2024 09:05 — 👍 1 🔁 0 💬 1 📌 0

By scholargoggler.com
22.11.2024 16:19 — 👍 1 🔁 0 💬 0 📌 0
.. by the way, I also created starter pack focused on DNA and RNA G-quadruplexes #G4 and included some buddies @sebastien-britton.bsky.social @granzhan.bsky.social @drzoewaller.bsky.social @paeschkelab.bsky.social @jschneek.bsky.social
Contact me if interested!
go.bsky.app/4VgiGa4
13.11.2024 21:50 — 👍 10 🔁 7 💬 4 📌 3
Created an #RNAbiology feed to talk RNA science! Let's use it.
@onamy.bsky.social @mgblango.bsky.social @graveley.bsky.social @nickingolia.bsky.social @hogglab.bsky.social @quaidmorris.bsky.social @snf.bsky.social @lucksmith.bsky.social
17.10.2023 12:23 — 👍 34 🔁 21 💬 9 📌 0
Widespread 3′UTR capped RNAs derive from G-rich regions in proximity to AGO2 binding sites
10/10
Our study opens new questions about these RNAs’ roles and functions. Dive into the full paper for all the details: rdcu.be/dZvPg
15.11.2024 17:14 — 👍 2 🔁 0 💬 0 📌 0

9/10
In summary, we demonstrate that that capped 3′UTR-derived RNAs arise from structured G-motif-rich regions bound by AGO2 and UPF1, and they localise separately from their parental mRNAs.
15.11.2024 17:14 — 👍 0 🔁 0 💬 1 📌 0

8/10
Using high-resolution imaging, we validated that 3′UTR-derived RNAs localise differently from their parental mRNAs within the cell, hinting at potential independent functions.
15.11.2024 17:12 — 👍 0 🔁 0 💬 1 📌 0
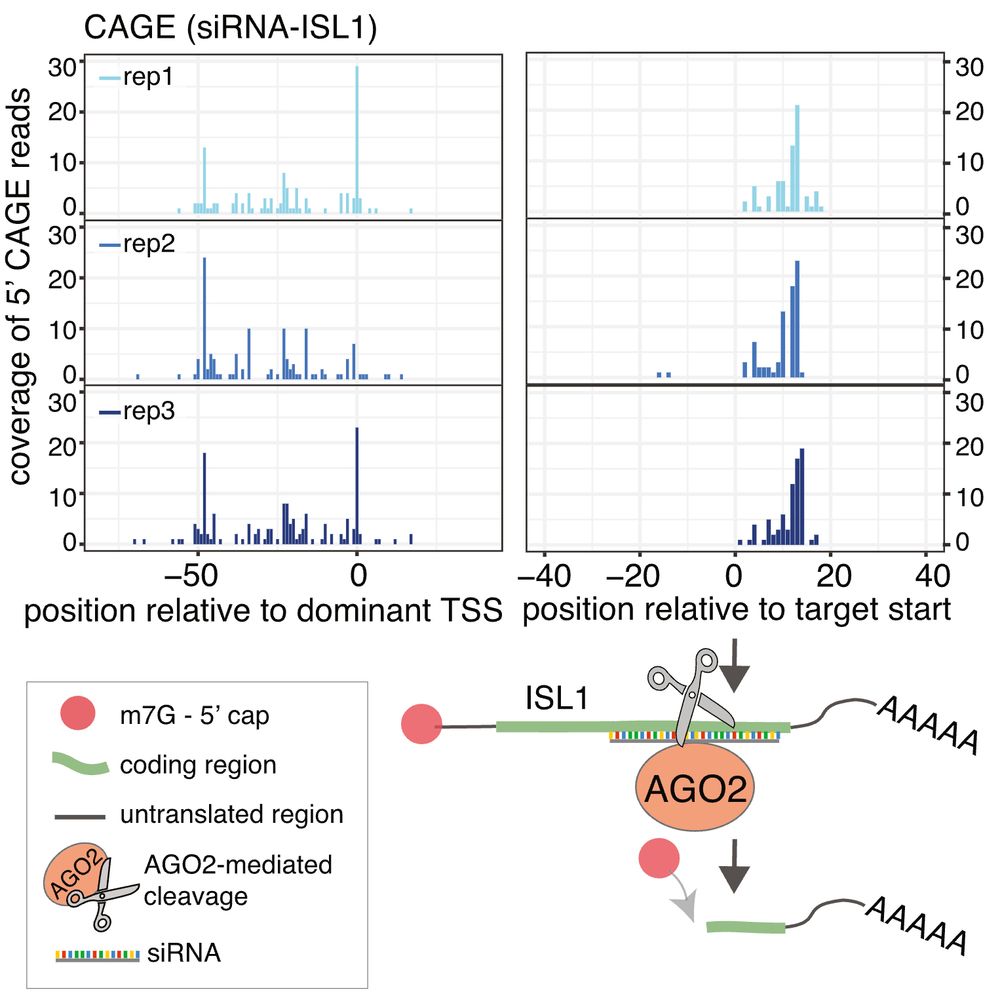
7/10
We also found that cytoplasmic capping can occur subsequent to AGO2/siRNA-mediated cleavage, by analysing FANTOM5 CAGE from siRNA-treated samples. Our findings demonstrate that capping can readily follow cytoplasmic cleavage, and is not exclusive to transcriptional initiation.
15.11.2024 17:11 — 👍 1 🔁 1 💬 1 📌 0
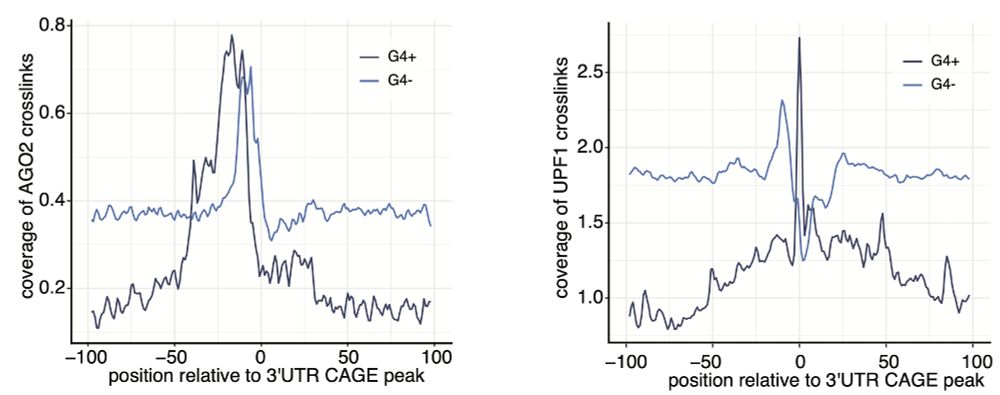
6/10
Our analysis shows that AGO2 binds upstream of 3′UTR-derived RNA start sites—independent of miRNA binding. Interestingly, G4 motifs influence the binding positions of both AGO2 and UPF1, shaping the capping sites.
15.11.2024 17:10 — 👍 0 🔁 0 💬 1 📌 0
Studying RNA decay at the NIH and Johns Hopkins University | KU Alum
We use human stem cell models to understand a range of neurodegenerative disorders, with particular focus on RNA metabolism and cellular autonomy. We are based at the National University of Singapore in the LSI: https://www.nus.edu.sg/lsi/
Social media account of the ERC SyG funded project UNLEASH. We will harness the splicing code for targeted control of gene expression using small molecules!
RNA, txtomics & the art of embracing variability
PI at the Centre for Cell Biology & @edinburgh-uni.bsky.social working on how cell size impacts cell functions | Organiser of Cell Size & Growth | Previously @Stanford, @crick.ac.uk & @Cambridge_Uni
Stadtman Principal Investigator @NIH / NIA. Computational Genomics, RNA, Aging, Stress Response, Senescence. Posts are my own.
Shy-guy-chris is interested in all sciency stuff, especially RNA biology and transcription. Currently working for Westicles at the University of Exeter.
Decoding how the gut thinks 🦠🪱🧠💪
Neuroscientist with interests in
#EnergyMetabolism #EntericNeurons #Fats
@crick.ac.uk @institutducerveau.bsky.social
Group leader in Bern, Cell-free translation enthusiast enjoying teaching, once upon @RNAKook on x-Twitter
https://scholar.google.com/citations?user=24B_5toAAAAJ&hl=en
https://www.ekarousis.net/
Since 1911, we have supported the molecular bioscience community, promoting the sharing of knowledge and enhancing career development.
Discover more about the Society: https://linktr.ee/biochemicalsociety
Studying RNA-binding proteins (RBPs) and how they modulate protein synthesis in normal and dysregulated cardiometabolic cells/tissues (and in other systems via collaboration). Especially interested in RBP regulation via intracellular signalling pathways.
Group leader and Wellcome Career Development Award fellow at the Department of Biochemistry, University of Cambridge. Interested in data-driven methods to understand gene regulation and genome organisation.
Basic Genomics specializes in advanced RNA sequencing technologies that enable precise detection and quantification of RNA molecules. Find out more at www.basic-genomics.com!
Group Leader @MRC LMS @Imperial College
Lab studying #redox #metabolism #ageing #Drosophila
Molecular evolution, chromatin, archaea and oddball biology. Associate Professor @oxfordbiochemistry.bsky.social Fellow @trinityoxford.bsky.social
A pioneer of sustainable open access, where research is always in progress.
RNA and ciliate molecular biologist, college prof, mom, living and working in the Pacific NW 👩🏻🔬🧬👩🏻🏫🔬
https://biology.wwu.edu/people/lees65
Postdoc in Bevilacqua Lab @PennState | RNA structure and ribozymes | Ph.D. UC Irvine, BS-MS IISER Bhopal (India)

















