
10 courses for my dream bioinformatics curriculum:
1. Unix Commands with Greg Wilson www.youtube.com/watch?v=U3i...
2. statistics and R with Rafael Irizarry rafalab.dfci.harvard.edu/pages/harva...
@hibernogruppe.bsky.social
Asst. Prof. JGU Mainz 🇩🇪 (he/him)| 🐝🧬 wild bee evolution | 🐟💻 salmonid genomics | https://github.com/joscolgan

10 courses for my dream bioinformatics curriculum:
1. Unix Commands with Greg Wilson www.youtube.com/watch?v=U3i...
2. statistics and R with Rafael Irizarry rafalab.dfci.harvard.edu/pages/harva...
We are thrilled to announce the first official release (v0.1.8) of #𝗯𝗲𝗱𝗱𝗲𝗿, the successor to one of our flagship tool, #𝗯𝗲𝗱𝘁𝗼𝗼𝗹𝘀! Based on ideas we conceived of long ago (!), this was achieved thanks to the dedication of Brent Pedersen.
1/n



📢 18-month PostDoc Fellowship – University of Montpellier (France)
🦠🦋 Interested in insect pathology, immunity, microbiome, or genomics?
We’re looking for a motivated candidate (<2 years post-PhD by mid-2026). 🧪
📩 Contact me ASAP for details!
⏳ Application deadline: 16/01/26
🔁 Please RT / Repost
PhD position available in evolutionary genomics/bioinformatics (hoehnalab.github.io/job_adverts/...). Topic: analyzing gene expression evolution across several firefly species and linking expression changes to genomic architecture. The position is jointly supervised with @anaevolcatalan.bsky.social
11.11.2025 09:00 — 👍 43 🔁 54 💬 4 📌 2🧬 PhD Position: Evolutionary & Comparative Genomics 🕷️
Join us at @uni-goettingen.de for a 3-year PhD (65% TV-L E13).
We will investigate the genomic & phenotypic impact of gene duplication across 233 arthropod genomes!
More infos 👇 and s.gwdg.de/eDrAAY
#Evolution #Genomics #Bioinformatics

Full paper now out in GENETICS:
The relationship between sexual dimorphism and intersex correlation: do models support intuition?
🔗 academic.oup.com/genetics/art...
#Evolution #QuantGenetics @GeneticsGSA

1/ a single job opening receives >1000 applications. (I am not kidding).
How to stand out?
Most bioinformatics CVs look the same: Python, R, RNA-seq, pipelines.
But hiring managers don’t care about skills on paper.
They care about proof. 🧵

FANTASIA's paper is finally out! Check it out if you're interested in alternative methods to homology for functional annotation in nonmodel organisms 🦐🐙🪱🧬🐝🪲
@gemmaeling.bsky.social @amrojasm.bsky.social @ibe-barcelona.bsky.social @cabd-upo-csic.bsky.social
@csic.es www.nature.com/articles/s42...
🧬🖥️ Thrilled that our paper "Tree-based differential testing using inferential uncertainty for RNA-seq" genome.cshlp.org/content/earl..., led by @noorpratap.bsky.social, & in collab with @mikelove.bsky.social, @fennecpaaaw.bsky.social and Jason Fan is now accepted at @genomeresearch.bsky.social. 1/4
21.08.2025 19:24 — 👍 39 🔁 9 💬 2 📌 1
Excited to share a new update to Mumemto, scaling MUM and conserved element finding to any size pangenome! Preprint out now w/ @benlangmead.bsky.social.
Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n
Massive thanks to @tegangaetano.bsky.social and @fmanfredini.bsky.social for impecably hosting while I was over - fantastic to hear about all the exciting research going on in Aberdeen🏴, meeting great people🍻, as well as seeing the wildlife🦭🦭🦭
23.05.2025 15:22 — 👍 2 🔁 0 💬 0 📌 0
Annotation of 200 insect genomes. Work driven by @stepbystepsolution.bsky.social and Mario Stanke within @gevol.bsky.social : www.biorxiv.org/content/10.1...
21.04.2025 05:37 — 👍 39 🔁 26 💬 1 📌 1
Simplify and elevate your data visualization with GGally, an R package designed to extend ggplot2.
Visualization: https://ggobi.github.io/ggally/
Details: https://statisticsglobe.com/online-course-data-visualization-ggplot2-r
#tidyverse #datavisualization #datasciencecourse #data #package
🐝Great to have this out @molbioevol.bsky.social and well done Hongfei Xu (not on BlueSky) for all the hard work
16.04.2025 15:53 — 👍 5 🔁 3 💬 0 📌 0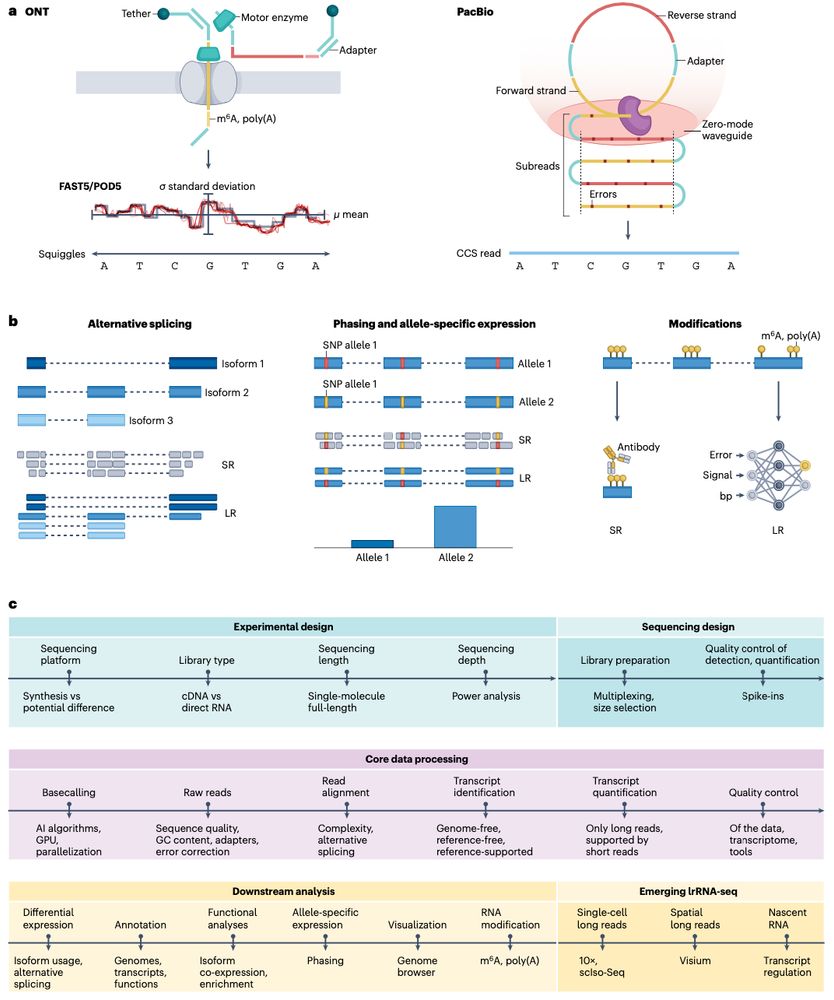

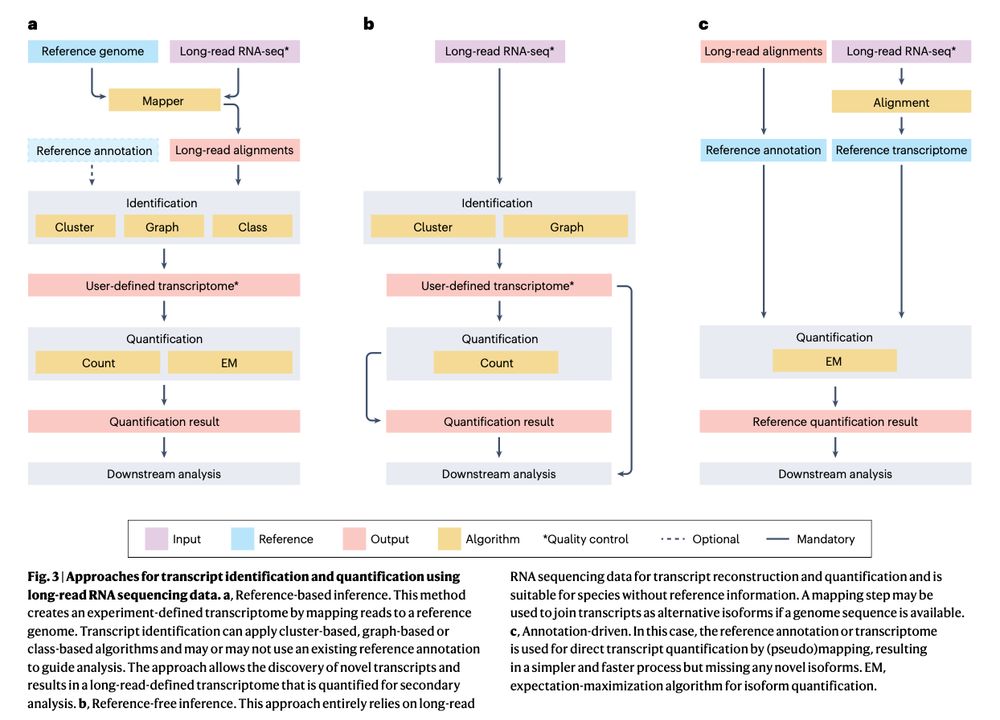
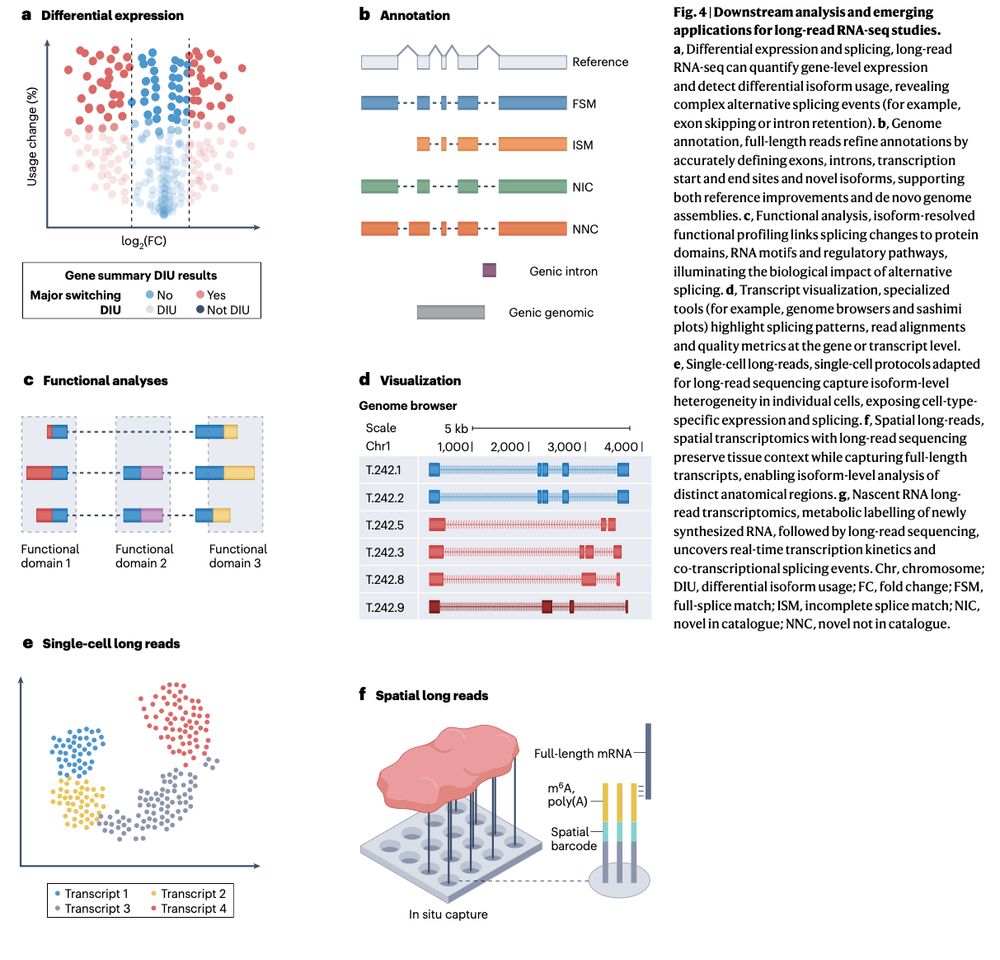
Review: Transcriptomics in the era of long-read sequencing www.nature.com/articles/s41... (read free: rdcu.be/efBjP) 🧬🖥️🧪
29.03.2025 08:57 — 👍 32 🔁 12 💬 1 📌 0
A local synteny visualization with 3 genes across 4 genotypes.
Using my Tidyverse local synteny visualization tools first time in 3 months. This a biosynthetic gene cluster with 2 P450s and a O-methyltransferase. Visualized across assemblies for 4 cultivars.
GitHub: github.com/cxli233/Tidy...
Our review on k-mers + pop gen is now published!
doi.org/10.1093/molb...
Friends, LiftOn is now published in @genomeresearch.bsky.social! LiftOn maps genes across genomes using a protein-centric, homology-based approach with both DNA & protein alignments
@stevensalzberg.bsky.social Mihaela Pertea, Jakob Heinz, Alaina Shumate, Celine Hoh, Alan Mao
doi.org/10.1101/gr.2...

Resulting from an @snsf-ch.bsky.social SPARK grant this took some time to mature, but the outcome is very imformative and builds a foundation for where to head next - how to liberate facts/information locked in the published literature www.biorxiv.org/content/10.1... #textmining #biodiversity
25.02.2025 19:31 — 👍 21 🔁 12 💬 1 📌 0Bitte am Sonntag wählen gehen! Noch nie war eine Wahl so wichtig für Deutschlands Zukunft.
#Btw2025

Wow. This is sooo good. Echoing @urbanevol.bsky.social this user’s manual for Ne should be required reading for popgen and conservation genetics folks. onlinelibrary.wiley.com/doi/10.1111/...
17.02.2025 18:26 — 👍 89 🔁 41 💬 1 📌 4
With @katiejenike.bsky.social and a bunch more of our fellow k-mer enthusiasts, we put together a manuscript on k-mers in biodiversity genomics. A guide if you will, that covers k-mers from basics to some really funky stuff...
genome.cshlp.org/content/35/2...

Finally ready to see the world! Theories of balancing selection - past, present and future: www.biorxiv.org/content/10.1... A true collaborative effort that brought together theoreticians and empiricists, models, data, and fresh perspectives on how balancing selection can shape genetic variation.
14.02.2025 16:49 — 👍 29 🔁 15 💬 2 📌 1
Olá, Brasil! 🇧🇷 Interested in a research stay in Germany? Are you currently a postdoc at a uni or research institution in Brazil? Our CAPES #Fellowship offers you funding for a #research stay for 6-24 mth! Join our info event on 18 Feb! www.humboldt-foundation.de/en/connect/i...
13.02.2025 15:08 — 👍 26 🔁 10 💬 0 📌 1
Principled PCA separates signal from noise in omics count data www.biorxiv.org/content/10....
13.02.2025 15:15 — 👍 12 🔁 2 💬 0 📌 0
ggalign: Bridging the Grammar of Graphics and Biological Multilayered Complexity 🧬🖥️🧪
Paper: https://www.biorxiv.org/content/10.1101/2025.02.06.636847v1
#Rstats Code: https://github.com/Yunuuuu/ggalign
Docs: https://yunuuuu.github.io/ggalign/
Book (Quarto): https://yunuuuu.github.io/ggalign-book/

New paper with Sarthak Mishra! Maybe it will bring you some joy given all this (waves arms around).
"Estimating recombination using only the allele frequency spectrum"
1/2
www.biorxiv.org/content/10.1...

What is a differentially expressed gene? https://www.biorxiv.org/content/10.1101/2025.01.31.635902v1 🧬🖥️🧪 https://github.com/Morris-Research-Group/bayexpress
05.02.2025 21:00 — 👍 38 🔁 11 💬 1 📌 0
A new tool, vcfsim, developed by Paimon Goulart, a stellar undergraduate researcher in my lab! vcfsim allows for the quick and easy simulation (via parameterized msprime simulations) of "all-sites" vcfs, with arbitrary ploidy and amounts of missing data.
www.biorxiv.org/content/10.1...


CASTER: Direct species tree inference from whole-genome alignments https://www.science.org/doi/10.1126/science.adk9688 🧬🖥️🧪 https://github.com/chaoszhang/ASTER
31.01.2025 21:00 — 👍 15 🔁 6 💬 0 📌 0