Thank you, Gerti!
28.02.2026 21:43 —
👍 0
🔁 0
💬 0
📌 0

Honoured to receive the Young Fluorescence Investigator Award at #BPS2026!
Huge thanks to the selection committee and to all the mentors and colleagues who supported me.
Excited to start new projects and keep using fluorescence to uncover molecular biology!
24.02.2026 17:51 —
👍 23
🔁 0
💬 1
📌 0

4Pi-SIMFLUX: 4Pi single-molecule localization microscopy with structured illumination - Nature Methods
4Pi-SIMFLUX is a single-molecule localization microscopy approach that achieves a near-isotropic resolution below 10 nm in whole mammalian cells.
We are happy to share our latest work, 4Pi-SIMFLUX, which combines structured illumination with interferometric detection to achieve near-isotropic 3D localization precision of 2–3 nm and resolve sub-10 nm structural features across whole mammalian cells.
www.nature.com/articles/s41...
rdcu.be/eQVxt
21.11.2025 03:00 —
👍 29
🔁 8
💬 0
📌 0
Congrats, Arindam!! Great news!! 🤩🥳
11.11.2025 22:25 —
👍 1
🔁 0
💬 1
📌 0

We're excited to announce the poster prize winners of #EESImaging 🔬
A special poster prize committee elected three winners:
Alara Kiris, @lumasullo.bsky.social and @wanluzhang.bsky.social. Congratulations!
Read on to learn about their research: s.embl.org/ees25-09post...
30.10.2025 12:21 —
👍 3
🔁 1
💬 0
📌 1
Thanks!!
20.10.2025 12:32 —
👍 0
🔁 0
💬 0
📌 0
Juliii ❤️ thanks for your kind words!! Looking forward to be part of the wonderful optics community in Argenitna!!
19.10.2025 02:14 —
👍 0
🔁 0
💬 0
📌 0

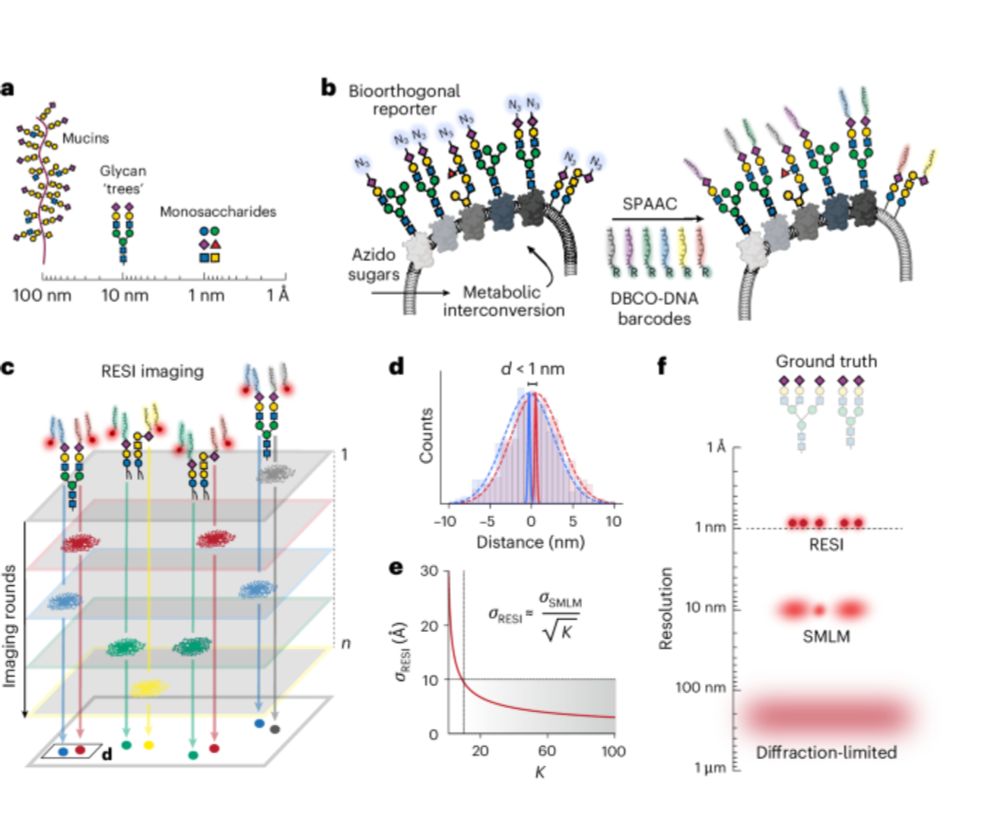
The image on the cover shows two sugars from the same cell-surface glycan separated by 9 Å, visualized with RESI (resolution enhancement by sequential imaging) enabled by metabolic labelling with DNA barcodes.
IMAGE: Luciano A. Masullo, Max Planck Institute of Biochemistry, Germany.
COVER DESIGN: Vanitha Selvarajan
Original paper: Masullo, L.A., et al. Ångström-resolution imaging of cell-surface glycans. Nat. Nanotechnol. 20, 1457–1463 (2025). https://doi.org/10.1038/s41565-025-01966-5
Abstract: Glycobiology is rooted in the study of monosaccharides, ångström-sized molecules that are the building blocks of glycosylation. Glycosylated biomolecules form the glycocalyx, a dense coat encasing every human cell with central relevance—among others—in immunology, oncology and virology. To understand glycosylation function, visualizing its molecular structure is fundamental. However, the ability to visualize the molecular architecture of the glycocalyx has remained challenging. Techniques such as mass spectrometry, electron microscopy and fluorescence microscopy lack the necessary cellular context, specificity and resolution. Here we combine resolution enhancement by sequential imaging with metabolic labelling, enabling the visualization of individual sugars within glycans on the cell surface, thus obtaining images of the glycocalyx with a spatial resolution down to 9 Å in an optical microscope.
Now online: October 2025 Issue.
- Focus Issue on #biosensing,
- DNA moiré superlattices,
- Sugars at Ångström-resolution,
- Solid-state #nanopores,
- Non-aqueous Li #batteries, -
- Neuromorphic vision,
- Peptide #hydrogels,
- Deep learning for #LNPs and more...
www.nature.com/nnano/volume...
17.10.2025 18:45 —
👍 8
🔁 5
💬 0
📌 0
Thread describing our work: bsky.app/profile/luma...
18.10.2025 14:33 —
👍 3
🔁 0
💬 0
📌 0

Our article "Ångström-resolution imaging of cell-surface glycans" made it to the cover of Nature Nanotechnology! 🤩 @natnano.nature.com
www.nature.com/nnano/volume...
18.10.2025 14:32 —
👍 74
🔁 8
💬 3
📌 0
Thank you, Inês! 😀
06.10.2025 07:02 —
👍 1
🔁 0
💬 0
📌 0
I am also very grateful to my previous mentors, Ralf Jungmann and Fernando D. Stefani, and many other amazing colleagues who have supported me throughout my career.
06.10.2025 06:24 —
👍 1
🔁 0
💬 0
📌 0
I am very grateful to Gabriel Rabinovich for his generous sponsorship and guidance throughout the application process, and I look forward to developing my research program within the stimulating environment of the Glycosciences Program at IBYME.
06.10.2025 06:24 —
👍 0
🔁 0
💬 1
📌 0
I’ll soon be looking for Master’s students, PhD students, and a postdoc to join the team—so if this sounds exciting to you, please don’t hesitate to reach out!
06.10.2025 06:24 —
👍 0
🔁 0
💬 1
📌 0
My research will focus on deciphering cell signaling at the molecular level by applying and developing cutting-edge super-resolution methods, with a particular emphasis on glycoimmune checkpoints, aiming to translate this knowledge into novel and improved therapies.
06.10.2025 06:24 —
👍 0
🔁 0
💬 1
📌 0
This grant gives me the opportunity to start building my own group, with support for setting up a state-of-the-art super-resolution microscopy lab.
06.10.2025 06:24 —
👍 1
🔁 0
💬 1
📌 0
Thrilled to announce some big news!
Excited to share that I’ve been awarded a Wellcome Trust Early Career Award to establish my research as a Group Leader in the Glycosciences Program at the Institute of Biology and Experimental Medicine (IBYME) in Buenos Aires, Argentina, beginning in early 2026.
06.10.2025 06:24 —
👍 55
🔁 4
💬 3
📌 1

Within this neuronal atlas we can reveal the three synapse classes, excitatory, inhibitory and the recently discovered mixed synapse. Organelle imaging of Peroxisomes (Pmp70) and the Golgi Apparatus (Golga5) reveals rare contact sides and even fused particles. (5/6)
02.10.2025 11:37 —
👍 7
🔁 1
💬 1
📌 0

To show the power of the technique, we acquired a 13-plex 200 x 200 µm2 neuronal atlas in 3D. With this atlas we map the interaction architecture of three neurons, resolving organelles, cytoskeleton, vesicles and synapses at single-protein resolution. (4/6)
02.10.2025 11:37 —
👍 5
🔁 1
💬 1
📌 0

We demonstrate speed-optimized left-handed DNA-PAINT by characterizing the sequence binding kinetics and resolving three main microscopy benchmarking targets, mitochondria, microtubules and nuclear pore complexes with <5 nm localization precision. (3/6)
02.10.2025 11:37 —
👍 7
🔁 1
💬 1
📌 0

The mirrored design of left-handed oligonucleotides allows the extension of the common 6 speed-sequences R1-R6 with their analogs L1-L6, enabling 12 target multiplexing with a standard secondary label-free DNA-PAINT workflow. (2/6)
02.10.2025 11:37 —
👍 6
🔁 1
💬 1
📌 0
Highly efficient 12-color multiplexing with speed-optimized DNA-PAINT. We are excited to share our latest paper in @natcomms.nature.com, using left-handed DNA to extend speed-optimized DNA-PAINT to 12 targets in a simple and straightforward way! 🧬👈🚀https://www.nature.com/articles/s41467-025-64228-x
02.10.2025 11:37 —
👍 28
🔁 9
💬 2
📌 2

One talk by Luciano Masullo at 10 am and another talk at 3 pm by Adam Cohen, both next Tuesday, October 7, at the MPI for Medical Research Heidelberg.
📣 📅 Invitation to two talks at our institute next Tuesday, October 7: 🤩 Luciano Masullo @lumasullo.bsky.social from @mpibiochem.bsky.social at 10 am, Adam Cohen @harvard.edu at 3pm, also online:
us06web.zoom.us/webinar/regi...
#maxplanck #mpimr #colloquium #medicalresearch #sciencetalks #science
02.10.2025 10:33 —
👍 2
🔁 1
💬 0
📌 0

Localization of single molecules with structured illumination and structured detection - Light: Science & Applications
Light: Science & Applications - Localization of single molecules with structured illumination and structured detection
Happy to share this article with my views on @elislenders.bsky.social and @vicidominilab.bsky.social work and discussing current developments in single-molecule localization combining structured excitation and detection. So many exciting perspectives in the field!
www.nature.com/articles/s41...
30.09.2025 18:17 —
👍 23
🔁 10
💬 2
📌 0
Congratulations, Rita! And thanks for all the amazing work at Nature Methods!
01.08.2025 23:21 —
👍 4
🔁 0
💬 1
📌 0
Thanks so much, Mahipal!!
30.07.2025 21:27 —
👍 0
🔁 0
💬 0
📌 0
Ångström-resolution imaging of cell-surface glycans
Now published in Nature Nanotechnology! 🤩
@natnano.nature.com
www.nature.com/articles/s41...
30.07.2025 14:58 —
👍 36
🔁 5
💬 1
📌 0