
Awesome to see @cfcollins.bsky.social first paper out from her PhD - a huge amount of work!!
Regulatory features determine the evolutionary fate of laterally acquired genes in plants url: academic.oup.com/mbe/article/...

Awesome to see @cfcollins.bsky.social first paper out from her PhD - a huge amount of work!!
Regulatory features determine the evolutionary fate of laterally acquired genes in plants url: academic.oup.com/mbe/article/...

Collins et al. combined expression, methylation, and genomic data to investigate long-term persistence of laterally acquired genes in the grass Alloteropsis, showing that those capable of regulating their own expression are more likely to persist.
🔗 doi.org/10.1093/molbev/msag042
#evobio #molbio
So cool. Multiple cell lineages contribute to the germline in maize -- and differences between apical and lateral branches! www.biorxiv.org/content/10.6...
09.02.2026 16:39 — 👍 14 🔁 5 💬 0 📌 1
Graphic announcing new AJB Synthesis article by Haley Branch.

Methodological illustration for asking whether X affects plants. “Question” provides an example of a question that both ecologists and evolutionary biologists ask, with categories of traits often of interest. “Experiment” shows trait examples and comparisons (populations, environmental, across time). Types of traits are where evolutionary biologists could expand their breadth. “Outcome” describes lasting effects of whether X affects traits to the point that it is passed on and contributes to future generations or it leads to extinction. “Outcomes” is where ecophysiologists could expand their work. Images courtesy of Dr. Stefanie Sultmanis (vein density), Dr. Beatrice Harrison Day (root section), and Dr. Vanessa Tonet (leaf cavitation).
🏵 Check out the latest #AJB Synthesis article, by Haley Branch! 🏵
The sleeping giant needs coffee: Overlooked areas for integrating plant #ecophysiology & evolutionary biology
doi.org/10.1002/ajb2...
#botany #plantscience #ecology #evolution

Updated tutorials for nQuack today. Want to predict ploidy level of sequence data? See this new tutorial on how to process and model data faster: mlgaynor.com/nQuack/artic...
14.10.2025 01:32 — 👍 14 🔁 11 💬 1 📌 0@oso-tee.bsky.social Congrats 🔥
16.09.2025 17:09 — 👍 1 🔁 0 💬 0 📌 0
Hybridization and introgression are major evolutionary processes. Since the 1940s, the prevailing view has been that they shape plants far more than animals. In our new study (www.science.org/doi/10.1126/...
), we find the opposite: animals exchange genes more, and for longer, than plants

Really excited to share our latest paper led by @simonaube.bsky.social. Fascinating results examining whether regulatory mutations can lead to adaptation as fast as coding mutations do. #mevosky #evobio #evoSky
www.biorxiv.org/content/10.1...

📣 The Call for Symposia for the Society for Molecular Biology and Evolution (SMBE) 2026 Annual Meeting is now open.
🌍 The meeting will be in Copenhagen June 28th to July 2nd
🖊️ We invite you to submit a symposium proposal and help shape the scientific content of SMBE 2026
Best the organisers

VpmaxA and Amax plotted by C4 biochemical subtypes (a and b, respectively), growth forms (c and d, respectively) and growth locations (e and f, respectively).
Environmental conditions drive variation in C4 photosynthetic capacity more than species traits
Fan, et al.
📖 nph.onlinelibrary.wiley.com/doi/10.1111/...
#PlantScience @yuzhenfan.bsky.social

Map of my no-fly itinerary from Italy to Bougainville, PNG
I've set off to Bougainville,PNG, for another #NoFly journey. I want to complete my research there. Travelling the ~25,000km distance from Italy by🚢,🚅,&🚌,I'll emit ~10 times less than✈️ (computations to come).Here the long read on my motivations: gianlu777.substack.c... 1/🧵
03.09.2025 08:01 — 👍 176 🔁 52 💬 10 📌 6
So excited to see our review on how to detect horizontal gene transfers and their impact on plant functional evolution finally published in @theplantcell.bsky.social!
Great teamwork between @lrsv-toulouse.bsky.social and LGDP-Perpignan 😎🥳🌱
academic.oup.com/plcell/advan...

Excited / nervous to share the “magnum opus” of my postdoc in Andreas Wagner’s lab!
"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)

Happy to see our latest work out in @molbioevol.bsky.social. We revisit the evolution of 5-methylcytosine across neglected eukaryotic supergroups, establishing an ancestral repressive role silencing genome invaders, both transposons and viral elements👾: academic.oup.com/mbe/advance-... 🧵 1/7
28.07.2025 10:03 — 👍 101 🔁 51 💬 6 📌 0
Our #research on #drought #recovery, now published with @springernature.com in @natcomms.nature.com:
Drought recovery in plants triggers a cell-state-specific immune activation.
doi.org/10.1038/s414...
Read thread below 👇
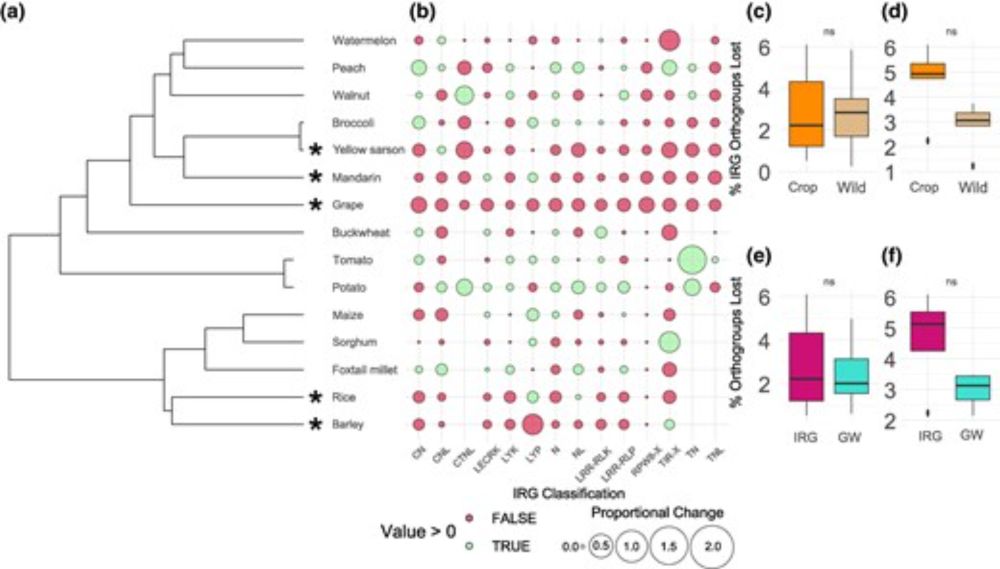
Bourne et al. analyzed immune receptor gene repertoires of 15 domesticated crop species, showing that grapes, mandarins, rice, barley, and yellow sarson exhibit reduced immune receptor gene repertoires compared to wild counterparts.
🔗 doi.org/10.1093/gbe/evaf147
#genome #evolution #PlantSky

The European Reference Genome Atlas (ERGA) is a pan-European community initiative to coordinate the generation of reference genomes representing European eukaryotic biodiversity @ergabiodiv.bsky.social #eseb2025
20.08.2025 09:06 — 👍 22 🔁 14 💬 1 📌 1
Out after peer review, collaborative study from Nordborg & Weigel labs with help from many others. Not the largest collection of new Arabidopsis thaliana genomes, but we hopefully put forward some good ideas for how to think about pangenomes and their analysis!
www.nature.com/articles/s41...
Today also, @thorstenreusch.bsky.social presented the fascinating work conducted on somatic mutations in the clonal seagrass Zostera! ✨ #ESEB2025
19.08.2025 14:24 — 👍 3 🔁 0 💬 0 📌 0Also appreciated the citation format used: Author, year, unique keyword. No journal name 🙌
19.08.2025 14:18 — 👍 1 🔁 0 💬 1 📌 0Inspiring talk from @marcrr.bsky.social presenting student Christabel Bucao’s work. Expression variability predicts retention and functional divergence of genes after WGD! « variability creates » #ESEB2025
19.08.2025 14:17 — 👍 4 🔁 1 💬 1 📌 0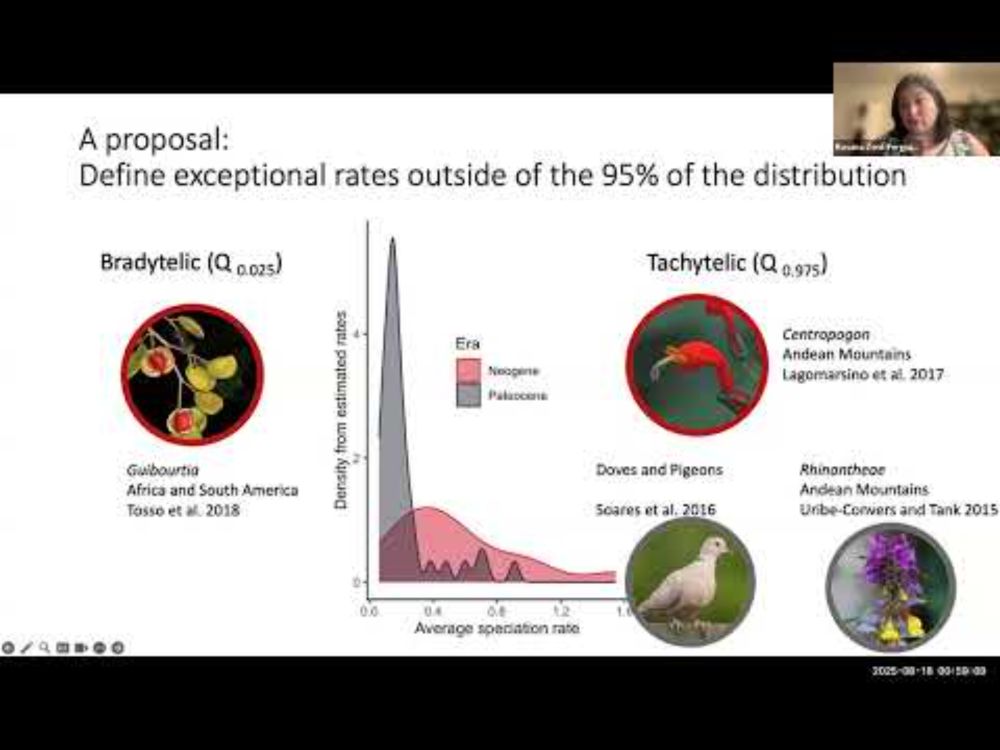
Unfortunately, I couldn't join in person #eseb2025, but the organizers kindly accommodated a video. Posted here in case you missed it!
youtu.be/n9Ls9-8p8x0?...
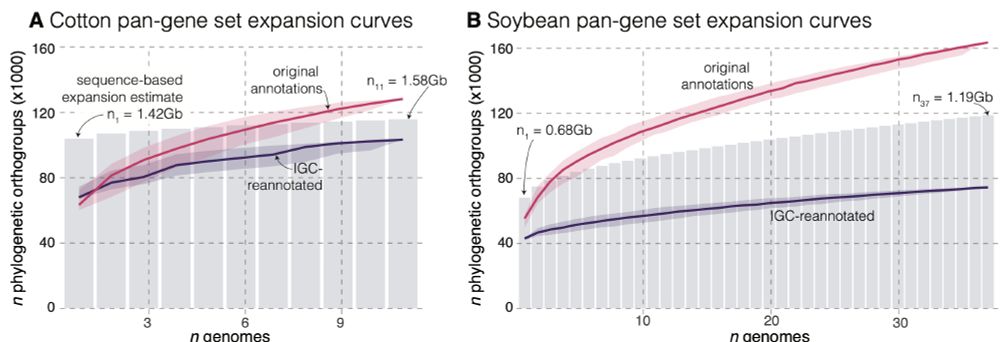
pangenome 'expansion' curves for cotton and soybean
Determining presence-absence variation (PAV) across reference genomes is a major goal of pangenome analysis. It turns out that A LOT of gene PAV is due to methodological artifacts.
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...

Picture of the Sagrada familia, symbol of Barcelona
Excited to be in Barcelona for #ESEB2025 So many great talks, familiar faces and cool posters already!
18.08.2025 17:54 — 👍 8 🔁 0 💬 0 📌 0This collaboration was so much fun!! I’m excited to see it online! www.cell.com/trends/genet...
09.08.2025 13:24 — 👍 30 🔁 10 💬 1 📌 1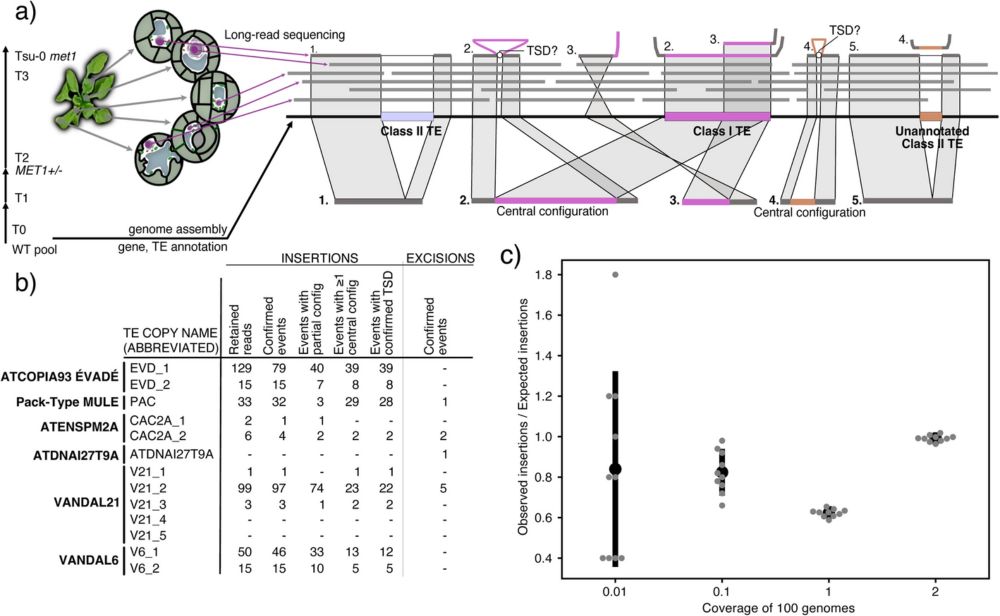
Paper led by @movillome.bsky.social on unbiased detection of (somatic) #TE insertions in #Arabidopsis with @pacbio.bsky.social long reads out after peer review. Thanks to academic editor Leandro Quadrana for shepherding it through the review process.
#plantscience
link.springer.com/article/10.1...?
Please RT!
I'm hiring a postdoc to work in my new lab at UCSB (!) on an NSF-funded project investigating the phenotypic consequences of WGD for photosynthesis and respiration.
Check out the full posting here:
recruit.ap.ucsb.edu/JPF02986
And more info about the lab here:
sharbroughlab.com



Il faut un sursaut collectif pour relancer l’action climatique et permettre l'accès à une transition pour tous : consolidation du cadre d’action publique, gouvernance solide, cap clair pour 2030 & 2040.
Dans notre rapport, nous formulons 74 recommandations : www.hautconseilclimat.fr/publications...
Could manage to add a bluesky widget on my website 🙌 ✨
👉 nicolasmouquet.free.fr
🤖 how to here : github.com/Vincenius/bs...
💡use you DID identifiers rather than your handle in the code (here is where to find it : ilo.so/bluesky-did/)
👏 thanks to @vincentwill.com for sharing 🧪
Qques mots forts que j'ai prononcés avec émotion @franceinfo.fr
C'est un mensonge de laisser croire que l'on s'adaptera sans casse irréversible dans une France à 4°C. On ne s'adapte pas a un effondrement de la biodiversité; on meurt avec!
Les régressions actuelles sont irresponsables & suicidaires.