Another hit from @alabamarivers.bsky.social and Southern Exposure
26.09.2025 13:42 — 👍 1 🔁 0 💬 0 📌 0

A two-panel Scooby-Doo meme. In the top panel, Fred pulls the mask off a villain labeled “Presence-absence-variation in plant genomes.” In the bottom panel, the unmasked villain is revealed to be “Annotation artifacts.”
🌿Plant pangenomes🌿 have a LOT of genomic presence-absence-variation.
Or do they???
A new @jgi.doe.gov preprint by @tomasbruna.bsky.social @jotlovell.bsky.social @avril-m-harder.bsky.social Avinash Sreedasyam takes a closer look
www.biorxiv.org/content/10.1...
18.08.2025 18:28 — 👍 21 🔁 8 💬 3 📌 0
'Hoods are out!!! Big thank you to @plantevolution.bsky.social for the support throughout. Luisa, @kdm9.bsky.social , @aconga.bsky.social , @hajkdrost.bsky.social it was intellectually stimulating ride with you, so congratulations!!!! #diversity #NLR #immune #pan-genome #graphs #networktheory
14.08.2025 20:22 — 👍 6 🔁 3 💬 0 📌 0

my new idea is to look at runs of homozygosity (ROH) only for mutations that are restricted to heterozygous state in other pops, suggesting they are low freq. and enriched for deleterious stuff. a good measure of inbreeding. I call my new statistic
12.08.2025 21:06 — 👍 65 🔁 6 💬 3 📌 1
This began as a @jgi.doe.gov Community Science Program (CSP) project in 2017 with the goal of uniting the diverse sorghum breeding and mapping populations in the framework of a 'pangenome'.
Thanks to a huge efforts across many stakeholders, the article was preprinted today. A 🧵 w/ what we found:
06.08.2025 20:32 — 👍 12 🔁 5 💬 1 📌 0
Postdoctoral Research Associate - Plant Systems Biology & The Center for Bioenergy Innovation (CBI)
Postdoctoral Research Associate - Plant Systems Biology & The Center for Bioenergy Innovation (CBI)
🚨 We're looking for a #postdoc to join our team at the Center for Bioenergy Innovation at Oak Ridge National Labs, TN 🚨
Project: Advance poplar bioenergy by understanding adaptive trait/genomic variation — breeding/pop gen/bioinformatics
Apply by 18-Aug
Apply here: jobs.ornl.gov/job/Oak-Ridg...
21.07.2025 13:52 — 👍 5 🔁 8 💬 0 📌 0
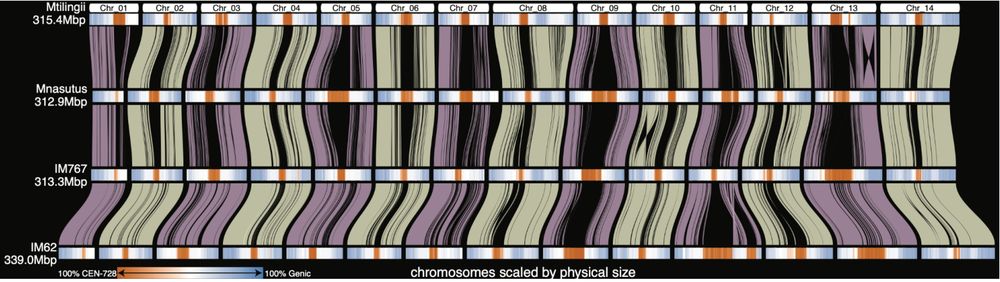
Fig. 3 | DEEPSPACE synteny map highlighting positions of centromeric repeats. Collinear blocks between the same chromosomes are shown as transparent ‘braids’ and chromosome segments are visualized as color-gradient rectangles along an x-axis that scales each genome by its physical size. Regions that do not map between chromosomes are visualized as black ‘wedges’ in the map. These can be due to ineffective unique mapping in highly repetitive centromeres (e.g. Chr 9 IM62-IM767), expansion of centromeric arrays (e.g. Chr 11 IM62-IM767) or sequence presence/absence. The orange-blue color gradient indicates regions that are gene-rich (blue) or centromeres (orange); fully saturated colors indicate that all sequence in those intervals is masked by that annotation type. White regions have neither genes nor Cent728 repeats, and are likely repeat-rich pericentromeres.
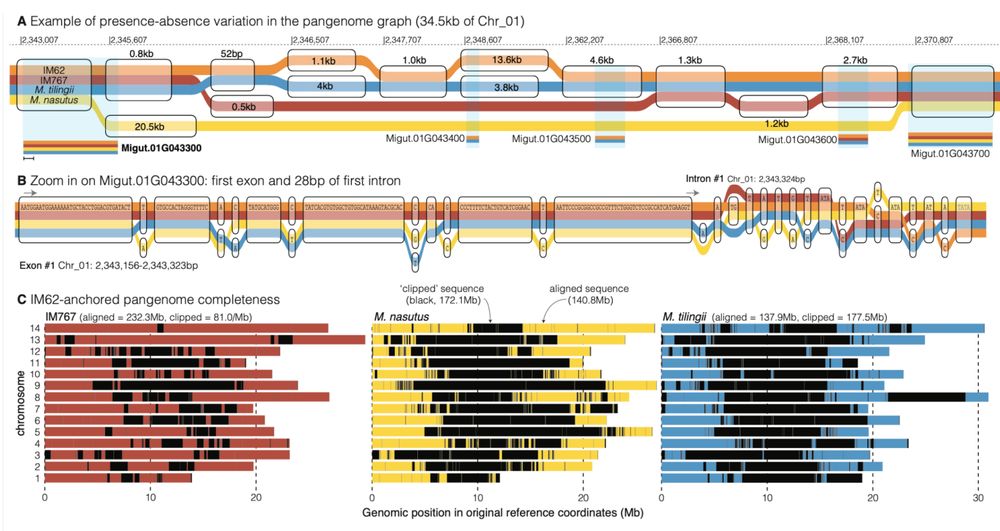
Fig. 4 | Exploration of the pan-genome graph. A 2.34-2.37Mb on chromosome 1 is a complex region in the genome that illustrates how the pan-genome graph handles large scale presence594 absence variation. Here, any variants smaller than 52bp are excluded so that the large insertion deletions in this region are apparent. Haplotypes containing similar sequences are binned in the transparent rounded rectangles (“nodes”), divergent but syntenic/orthologous sequences have stacked nodes, and deleted sequences show the path outside of a node. B The first exon and the first 28bp of the first intron of a gene with family members across all four genomes is shown as an example of how SNPs and INDELs appear at the base pair level in a sequence graph. C The positions of aligned sequence retained in the graph (colored following the tubes in panels A-B) and that which is un-alignable and clipped (black) are presented in a pan-genome anchored by IM62.
Ok, fine I'm willing to admit it now: Mimulus is interesting www.authorea.com/doi/full/10....
10.06.2025 22:15 — 👍 45 🔁 14 💬 4 📌 0
That time in 2013 when this crabeater seal decided to see what this noisy orange menace was doing in its territory....In 2013 near Peter I Island in #Antarctica on our research cruise (LMG 13-12). Bonus (the 'wait for it' at the end): amazing sunset/sunrise around 1am that night :) 🧪🌎🦑🐧🇦🇶
22.05.2025 11:22 — 👍 42 🔁 9 💬 1 📌 0
I remember this like it was yesterday 😄
22.05.2025 22:05 — 👍 1 🔁 0 💬 1 📌 0
Such a cool talk on a sweet system! Looking forward to seeing what else you find 🦎
19.05.2025 20:27 — 👍 1 🔁 0 💬 0 📌 0
Wow, the cover looks great!
Nice work Patrice and @roederlab.bsky.social
The GENESPACE plot uses our new @jgi.doe.gov Pennycress and Brassica rapa genomes built in collaboration with @spicybotrytis.bsky.social & Katie Greenham, hosted on phytozome
14.05.2025 13:39 — 👍 8 🔁 3 💬 0 📌 0
GitHub - yangao07/longcallD: A local-haplotagging-based small and structural variant caller
A local-haplotagging-based small and structural variant caller - yangao07/longcallD
longcallD is a new variant caller for genomic long reads. It jointly calls phased small and structural variants. Single binary, one command line for the whole process. Comparable accuracy to mainstream callers. Great work by Yan Gao. github.com/yangao07/lon...
24.03.2025 16:53 — 👍 105 🔁 49 💬 3 📌 3
Post a warning.
24.03.2025 17:32 — 👍 1 🔁 0 💬 0 📌 0
"Our results reveal substantial differences between pipelines, with many inversions either misrepresented or lost. Most notably, recovery rates remain strikingly low, even with the most simple simulated genome sets, highlighting major challenges in analyzing inversions in pangenomic approaches."
18.03.2025 16:53 — 👍 8 🔁 3 💬 0 📌 0
Our paper on the past, present, and genomics-enabled future of American chestnut (Castanea dentata) tree breeding for resistance to invasive pathogens is on BioRxiv.
🧵with some background and a few things we found 👇 bsky.app/profile/bior...
03.02.2025 21:23 — 👍 29 🔁 12 💬 1 📌 0

VG logo
A new version of our variation graphs (vg) tool, "Boccaleone," is now available! Updates include:
🔵 HiFi and Oxford Nanopore read support to the Giraffe read mapper
🔵 Faster read sorting
🔵 New Unix manual page
Link: github.com/vgteam/vg/re...
29.01.2025 17:18 — 👍 8 🔁 4 💬 1 📌 0

Are you headed to #PAG32 and interested in plant reproduction? 🌼 Come check out my talk in the “Sex Chromosomes and Sex Determination” session on Saturday!
I’ll be talking about ancient ZW sex chromosomes, the impact of polyploidy on sexual system evolution, “leaky” dioecy, and much more.
08.01.2025 14:02 — 👍 24 🔁 4 💬 0 📌 0

A) Summary of Camelina sativa progenitors and allopolyploid formation. B) Synteny between Arabidopsis thaliana and the three Camelina sativa subgenomes
What's better than one polyploid genome? Twelve polyploid genomes! We report a pangenome of the hexaploid oilseed crop Camelina sativa and investigate pangenomic variation and genome evolution in a complex allopolyploid genome. www.biorxiv.org/content/10.1...
17.08.2024 17:30 — 👍 23 🔁 14 💬 3 📌 1

Comparative genomics plot of sugarcane
Our groups' goal is to produce genomes that are useful; this does not always mean 'telomere-to-telomere' assemblies. The hybrid sugarcane genome is a nice example of this. The article is out this week in Nature: www.nature.com/articles/s41...
29.03.2024 17:14 — 👍 26 🔁 11 💬 1 📌 0

GenotypeGVCFs and the death of the dot
If you have been keeping up with our GATK release notes, then you know that we have been rolling out a number of backend changes, tools, and features to GATK that we are hoping will improve the eff...
@ksamuk.bsky.social pointed this out on the other site, but I don't see much discussion here.
GATK will now encode missing genotypes as "0/0", or homozygous ref, instead of "./." in VCF files. To identify sites with missing data, you'll have to check the DP field.
07.02.2024 15:18 — 👍 10 🔁 16 💬 5 📌 1

HudsonAlpha - Staff Scientist - Genome Sequencing Center - Huntsville, AL
Position: STAFF-SCIENTIST: Evolutionary gen
🚨We're hiring🚨
We are looking for a scientist to bridge the gap between genomics and breeding, focusing on traditionally under-invested crops and regions. It's physically located at Colorado State University in beautiful Ft. Collins, CO.
Pls share!
hudsonalpha.applicantpro.com/jobs/3211389
30.01.2024 17:37 — 👍 21 🔁 28 💬 2 📌 1
A new study shows that flowers have evolved over the past three decades to have less sex—potentially in response to the disappearance of bees. It could be a death spiral for some species. Here’s my story. 🧪 Gift link: www.nytimes.com/2024/01/04/s...
04.01.2024 14:34 — 👍 90 🔁 42 💬 7 📌 11
Alabama Rivers Alliance is a statewide network of groups working to protect and restore all of Alabama’s water resources through building partnerships, empowering communities & advocating for sound water policy & its enforcement. #DefendRivers #WaterIsLife
Fish biodiversity, genomics. Illinois Natural History Survey Asst Research Scientist. Also aquarium fish hobbyist and plant parent. Profile pic: With a tamandua knifefish. He/him
Assistant Professor @uarizona; macro-evolution, data science, and some ecology; Lab website: https://datadiversitylab.github.io/; Blog: https://ghost.cromanpa.synology.me/
RepeatMasker is a tool that annotates transposable elements, satellites and tandem repeats in genomic sequences.
Evolutionary biologist. Research Leader at Royal Botanic Gardens, Kew.
Genome Data Scientist @ DOE Joint Genome Institute
Our Mission: To improve the human condition through plant science 🌱
Located in St. Louis, Missouri, the Donald Danforth Plant Science Center is the largest independent non-profit institute dedicated to plant science in the world.
Assistant Professor - Mississippi State University - evolutionary biologist - Penstemon
Website: benstemon.github.io
Botanist. University of Georgia. Plant evolutionary biology, phylogenomics, comparative genomics.
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
lover of birds, nature, evolutionary biology, diversity in STEM, peace and good will
~ Unf*ckwithable ~ espnW, Host of “Good Game with Sarah Spain” on iHeart, Emmy- & Peabody-Winning Journalist, Washed-up Heptathlete, Cornell grad, Chicago gal.
Pre-order my new book “Runs In The Family” - www.tinyurl.com/RITFbook
talkin' hoops 🏀🎙️ @tribeathletics | producer @wmnmedia Good Game with Sarah Spain 🍊| @wmtribewbb 💚 & @cronkite_asu 🔱 alum | @risingmstars class of ‘24 | #HOOPER‼️
A lover of chromosome biology 🧬🔬🔎 Studying (not only) meiosis, centromeres & genome evolution in plants!
Group leader at MPI for Plant Breeding Research in Cologne, Germany
Lab website:
https://www.mpipz.mpg.de/5028512/marques
Tree breeder 🧬🌳🍒
Beekeeper 🐝🍯🐝
Opinions expressed here are representative of Doc Rhoades Honey Company and no other entities
she/her
https://kathleenrhoades.com/
The Vertebrate Genomes Project (VGP) aims to generate error-free genome assemblies for all 70,000 vertebrate species, offering a powerful resource to advance research in biology, genomics, conservation, medicine, and bioinformatics.
On the academic job market | How are species compared to one another across different genomic regions? Postdoc at Langmead Lab, Johns Hopkins | Comparative #genomics at scale | Formerly at UNIL/SIB/WUR | sinamajidian.github.io














