PREreview of “Molecular Drivers of RNA Phase Separation”
Authored by Giovanni Bussi and 1 other author
New #openreview from our group at prereview.org/reviews/1672...! @sdimarco.bsky.social and @bussigio.bsky.social reviewed a #preprint by @potoyan.bsky.social on the Molecular Drivers of #RNA Phase Separation. Also available at doi.org/10.5281/zeno...
02.08.2025 15:19 — 👍 1 🔁 1 💬 0 📌 0
Our research presented today at the Telluride meeting on #RNA dynamics! @bussigio.bsky.social will report on some of our ongoing projects: the recent #CASP16 solvation challenge, constant pH #MD simulations on #RNA, and 2D ensemble reconstructions using #DMS data
22.07.2025 14:21 — 👍 3 🔁 0 💬 0 📌 0
A review reporting on the discussions held at the recent @cecamevents.bsky.social workshop on enzymatic plastic degradation that we co-organized in Trieste is now available on #arxiv
22.07.2025 14:18 — 👍 2 🔁 1 💬 0 📌 0
Cambridge Ellis Unit Summer School on Probabilistic Machine Learning 2025 – ELLIS Cambridge Unit
Our research presented at the Cambridge Ellis Unit Summer School on Probabilistic Machine Learning! Giuseppe Sacco will present a poster reporting our machine-learned model for DMS-guided ensemble prediction of RNA systems www.ellis.eng.cam.ac.uk/summer-schoo...
16.07.2025 12:49 — 👍 3 🔁 1 💬 0 📌 0
The review has been already submitted to a journal — but we’re committed to improving it. Comments, suggestions, and discussions are very welcome! 🙏
14.07.2025 06:50 — 👍 2 🔁 1 💬 0 📌 0
We cover recent advances in atomistic simulations of #RNA: from #folding and #ion binding to interactions with #proteins and #ligands. A broad overview, with many references and open questions.
14.07.2025 06:50 — 👍 1 🔁 1 💬 1 📌 0
StatPhys29 – 2025 Florence, Italy – The 29th International Conference on Statistical Physics
Our research presented at #StatPhys29! Ivan Gilardoni will present a poster reporting on our methodological work in integrating molecular simulations and experimental data statphys29.org
14.07.2025 06:47 — 👍 2 🔁 1 💬 0 📌 0
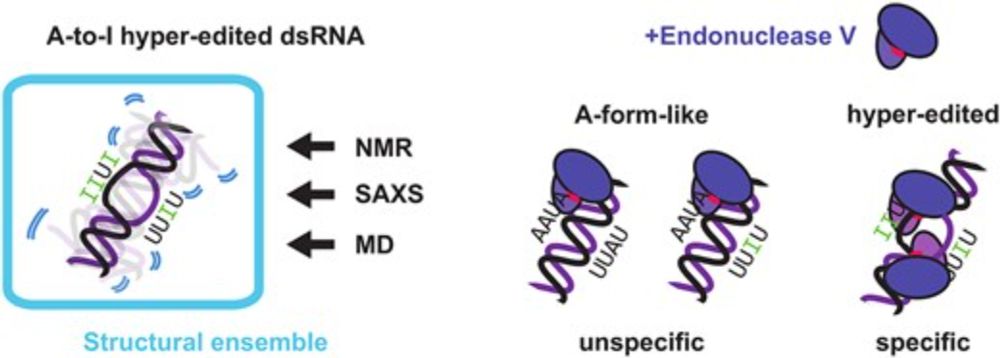
Unique conformational dynamics and protein recognition of A-to-I hyper-edited dsRNA
Abstract. Adenosine-to-inosine (A-to-I) editing is a highly abundant modification of double-stranded RNA (dsRNA) and plays an important role in posttranscr
📢 Paper with @sattler-lab.bsky.social now published in @narjournal.bsky.social ! doi.org/10.1093/nar/... We combine #NMR, #SAXS, and #MD to construct conformational ensembles, showing that A-to-I hyper-editing leads to significant structural dynamics. MD simulations by @piompons.bsky.social
26.06.2025 14:36 — 👍 12 🔁 6 💬 0 📌 0
A paper from our group (doi.org/10.1093/narg...) has been tested and reviewed in ReproHack www.reprohack.org/review/151! We got interesting feedback on how to improve documentation of our notebooks. Very useful initiative!
26.06.2025 05:48 — 👍 3 🔁 0 💬 0 📌 0
PREreview of “Minimum-Excess-Work Guidance”
Authored by Giovanni Bussi and Olivier Languin-Cattoën
New #openreview from our group at prereview.org/reviews/1573...! @bussigio.bsky.social and @ollyfutur.bsky.social reviewed a #preprint by Simon Olsson showing their Minimum-Excess-Work Guidance method. Also available at doi.org/10.5281/zeno...
25.06.2025 08:15 — 👍 5 🔁 0 💬 1 📌 0
Protein Electrostatics 2025, Lisbon
Our research presented at the proteinelectrostatics.org Protein Electrostatics 2025, Lisbon! @tf-silva.bsky.social shared our constant pH simulations on #RNA systems, from oligomers to introns!
23.06.2025 17:56 — 👍 4 🔁 1 💬 0 📌 0
Our research presented at the meeting of the Division of Chemistry of Biological Systems of the @socchimita.bsky.social! Elisa Posani will show our integrative modelling of #RNA using #cryoEM data and @sdimarco.bsky.social will share our simulations of #RNA membrane interactions
18.06.2025 09:13 — 👍 1 🔁 1 💬 0 📌 0
Our research presented at the GRC on computational #NMR organized by @katjapetzold.bsky.social in Les Diablerets! @bussigio.bsky.social will share our integrative approach to reconstruct the impact of A-to-I modifications on dynamics and our protocol to predict Mg2+ binding sites in #RNA molecules
04.06.2025 07:22 — 👍 6 🔁 2 💬 0 📌 0
Evaluation of novel computational methods to identify RNA-binding protein footprints from structural data
A monthly journal publishing high-quality, peer-reviewed research on all topics related to RNA and its metabolism in all organisms
📢 Paper on the RBP Footprint Grand Challenge—launched at RNA2021 — is out in @rnajournal.bsky.social doi.org/10.1261/rna....! A community-driven benchmark of methods for predicting RBP binding sites, with contributions from @tfroehlking.bsky.social, Mattia Bernetti, and @bussigio.bsky.social
27.05.2025 05:37 — 👍 10 🔁 2 💬 0 📌 0
Congratulations to @tf-silva.bsky.social for winning the best poster prize at the #RNA modelling across scales @cecamevents.bsky.social workshop, describing our constant pH simulations of RNA systems 👏
21.05.2025 18:21 — 👍 5 🔁 1 💬 0 📌 0
Our research was featured at the @cecamevents.bsky.social meeting #RNA modelling across scales at SISSA, Trieste! @ollyfutur.bsky.social gave a talk on our enhanced sampling strategy for exploring RNA-Mg2+ binding, and all other group members presented posters on their research
21.05.2025 18:19 — 👍 6 🔁 1 💬 0 📌 1
Our integrative approach using #MD and #cryoEM data to construct structural ensembles of #RNA just published on @natcomms.bsky.social doi.org/10.1038/s414... Lead by Elisa Posani, with @magistratolab.bsky.social @bonomimax.bsky.social @pjanos.bsky.social @navtejtoor.bsky.social and Daniel Haack 🎉
16.05.2025 09:16 — 👍 27 🔁 8 💬 0 📌 0
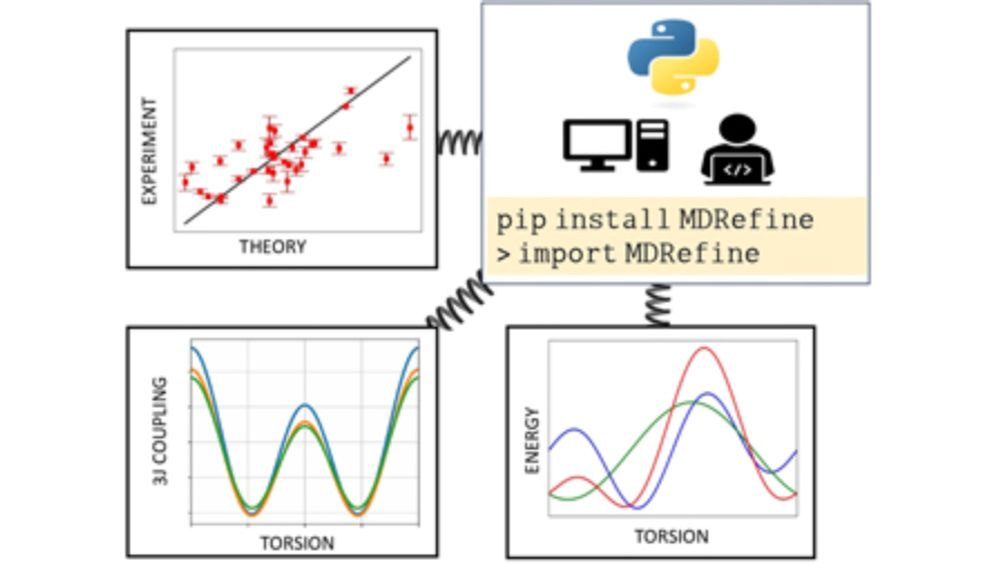
GitHub - bussilab/MDRefine
Contribute to bussilab/MDRefine development by creating an account on GitHub.
Work done by Ivan Gilardoni, with help from @piompons.bsky.social @tfroehlking.bsky.social and @bussigio.bsky.social Code is available on GitHub github.com/bussilab/MDR... . Play with it using `pip install MDRefine` pypi.org/project/MDRe...
15.05.2025 12:55 — 👍 5 🔁 2 💬 0 📌 0
📢 Ten days left to apply for a #PhD in Molecular and Statistical Biophysics at SISSA 👇 Deadline is April 28!
17.04.2025 18:40 — 👍 4 🔁 1 💬 0 📌 0
🚨 New paper! By combining protein language models, clustering, and DCA, we reveal how MSA subclusters drive AF2 predictions toward alternative conformations, and how statistics from the clustered sequences can inform deisgn of mutations that shift conformer populations.
16.04.2025 14:34 — 👍 10 🔁 4 💬 0 📌 0
Join us – Molecular and Statistical Biophysics
📢 Apply for the #PhD 🎓 in Molecular and Statistical Biophysics at SISSA in lovely Trieste! 4 fully funded fellowships starting Oct 2025. ~8 months of classes, then choose your project. Remote entrance exam. Application deadline: 28.4.2025. Please repost! 👉 msb.sissa.it/join-us/ #joinus
08.04.2025 17:26 — 👍 2 🔁 3 💬 0 📌 1
Our community letter on #FAIR principles in #moleculardynamics simulations has been published on @naturemethods.bsky.social 🎉
05.04.2025 09:56 — 👍 17 🔁 4 💬 0 📌 0
Our research presented at an @ebi.embl.org workshop on Computational approaches to predict small-molecule/RNA interaction! Today @bussigio.bsky.social will show our #CASP16 results on predicting Mg binding sites and our attempts to model chemical probing experiments
02.04.2025 06:37 — 👍 10 🔁 1 💬 0 📌 0
We are sill accepting applications for poster presentations at the CECAM conference 'RNA Modelling Across Scales". Join us in the beautiful Trieste ☀️🏖️🏰! Apply now: www.cecam.org/workshop-det... Co-organized with @bussigio.bsky.social @marcodevivo.bsky.social
27.03.2025 19:04 — 👍 10 🔁 5 💬 0 📌 0
We are finalizing the program of the @cecamevents.bsky.social RNA modelling across scales www.cecam.org/workshop-det... at SISSA, Trieste, Italy, May 19-22. Please note that we still have a few slots for in person attendance. We will accept applications for the next few weeks, until fully booked!
27.03.2025 17:15 — 👍 11 🔁 7 💬 0 📌 0
Still a few days remaining to apply for the RNA modelling across scales @cecamevents.bsky.social workshop at SISSA. Deadline March 23, hurry up!!
20.03.2025 16:55 — 👍 5 🔁 2 💬 0 📌 1
PhD student in Statistical Biophysics at SISSA, Trieste, Italy
Computational perspective on molecular evolution & function @areasciencepark
molecular simulations lab in between ICM and IQAC at @CSIC.es | investigating the molecular dimension of life, one atom at the time | don't follow us, we are lost too | led by F Colizzi
PostDoc, @gervasiolab.bsky.social UNIGE |
PhD, @bussilab.bsky.social SISSA |
#machinelearning, #RNA, #Proteins, #moleculardynamics matching experiments
Molecular simulations and AI at Area Science Park
Full professor at SISSA, msb.sissa.it group | Group leader @bussilab.org | Founder and developer @plumed.org
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app