
Our study is now published on JCIM🎉
We expanded and refined the preprint thanks to the insightful feedback from reviewers!
paper: pubs.acs.org/doi/10.1021/...
code: github.com/RitAreaScien...
@piompons.bsky.social
Molecular simulations and AI at Area Science Park

Our study is now published on JCIM🎉
We expanded and refined the preprint thanks to the insightful feedback from reviewers!
paper: pubs.acs.org/doi/10.1021/...
code: github.com/RitAreaScien...

Glad I had the chance to present our recent work in Trento at the conference "Molecular Biophysics at the Transition State"
08.07.2025 10:02 — 👍 3 🔁 0 💬 0 📌 0
Workshop not to be missed at Area Science Park (Trieste) on the 9th of September.
Special Guests: David Robertson (UK), @grovearmada.bsky.social (UK), Emanuele Andreano (IT)
www.areasciencepark.it/en/events/ai...
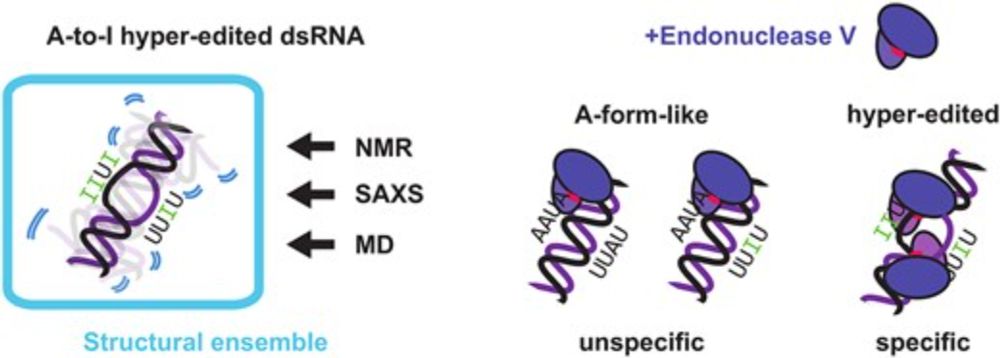
📢 Paper with @sattler-lab.bsky.social now published in @narjournal.bsky.social ! doi.org/10.1093/nar/... We combine #NMR, #SAXS, and #MD to construct conformational ensembles, showing that A-to-I hyper-editing leads to significant structural dynamics. MD simulations by @piompons.bsky.social
26.06.2025 14:36 — 👍 13 🔁 6 💬 0 📌 0🔥 Two PhD positions open @UniTrieste funded by @AreaSciencePark! 🔥
Join the Laboratory of Data Engineering to advance research in AI and its scientific applications.
We’re looking for motivated students ready to dive into interdisciplinary research in deep learning and AI.

Almost at the end of the exciting @cecamevents.bsky.social conference 'RNA Modelling across scales' at SISSA, Trieste, Italy! 🎉 Fantastic speakers and great science. Organized with @bussigio.bsky.social @marcodevivo.bsky.social #RNA
21.05.2025 19:22 — 👍 19 🔁 8 💬 0 📌 3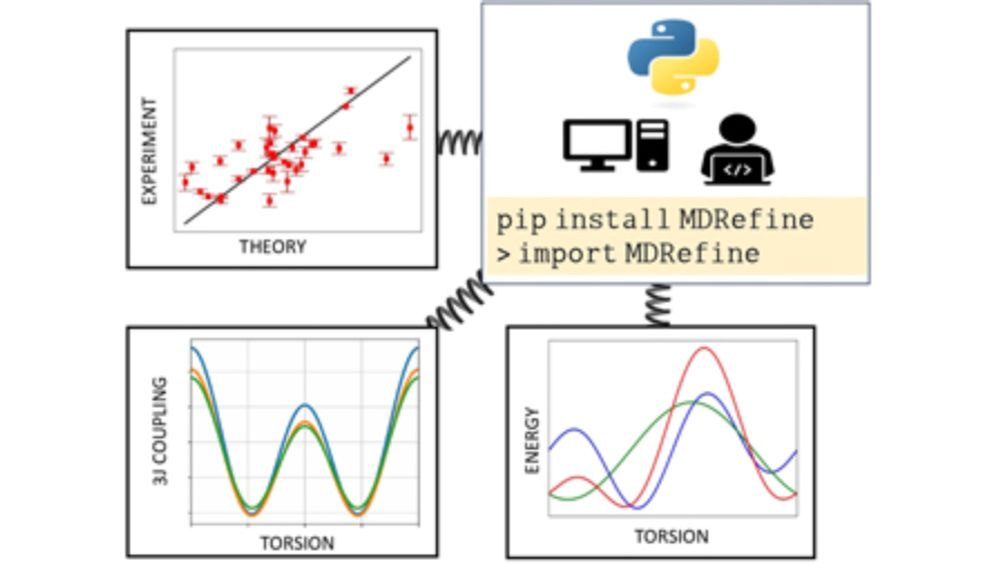
The manuscript describing MDRefine, our #python package for integrating experimental data and #moleculardynamics trajectories using reweighting methods, has been published on J Chem Phys doi.org/10.1063/5.02...!
15.05.2025 12:53 — 👍 12 🔁 2 💬 2 📌 0🚨 New paper! By combining protein language models, clustering, and DCA, we reveal how MSA subclusters drive AF2 predictions toward alternative conformations, and how statistics from the clustered sequences can inform deisgn of mutations that shift conformer populations.
16.04.2025 14:34 — 👍 10 🔁 4 💬 0 📌 0Our research presented today at AreaSciencePark! @bussigio.bsky.social will discuss our recent results in enhanced sampling of #RNA Mg interactions, integration of #cryoEM data, and refinement of force fields for modified nucleotides. www.areasciencepark.it/en/scientifi...
22.01.2025 06:46 — 👍 7 🔁 2 💬 0 📌 1