

Today we celebrated the "graduation" of our UU_plants
Rclub 2024! We hope to keep empowering young researchers to improve their R skills!
Special highlight for the great cake made by PhD candidate Dani (bottom left)!
#Rstudio #tidyverse
@leonardojo.bsky.social
Postdoc fellow at Wageningen University Gene regulatory mechanisms & plant development. @ucdavis Alumni First Gen. He/Him.


Today we celebrated the "graduation" of our UU_plants
Rclub 2024! We hope to keep empowering young researchers to improve their R skills!
Special highlight for the great cake made by PhD candidate Dani (bottom left)!
#Rstudio #tidyverse

It took me SEVEN years to figure out that you can save a ggplot as a vectorized .svg file ggsave("file.svg"). The svg file can be opened on pptx/ai/inkspace and it will be completely vectorized, even text are still recognized as text boxes! No more recreating plots on illustrator!!!!
28.08.2024 15:45 — 👍 51 🔁 13 💬 2 📌 1
Very happy to share this review with @mnodine1
www.sciencedirect.com/science/arti...
Here, we describe what is known (or unknown) about the mechanisms of chromatin reprogramming during the early embryogenesis in plants.
A lot of exciting questions that still need to be explored.
#plantbiology

Bouquet of flowers and a framed certificate for the best student evaluations
Our course Plant Development & Environment 🌱 was the highest rated level 3 in BSc Biology!
I love the enthusiastic 🤩 students & the excellent teachers 🙏@daanweits @leonardojo.bsky.social @andruromanowski.bsky.social @dkawa.bsky.social @rsasidharan.bsky.social @viktoriiavoloboeva.bsky.social
I'm relatively new to building shiny apps, so any comments and suggestions would be greatly appreciated!
You can open a ticket on github or send me an email: l.jo@uu.nl
Thanks for the help!
It's really raw, but I would like to ask people to try this out on their own ggPlantmaps to see potential errors that can occur.
A link for the beta test instructions:
github.com/leonardojo/g...


[Beta test] ggPlantmap + shinyapp.
Seeking help from the community:
I created a function that allows users to create interactive shiny apps to navigate through multiple values (ex: genes) on top of their #ggPlantmap.

OMG! My great mentor John Harada is the recipient of the @aspbofficial.bsky.social Stephen Hales award 2024! This is HUUUGE! He just entered the plant biology hall of fame! Im so happy for him!
11.04.2024 21:31 — 👍 5 🔁 0 💬 0 📌 0
The EPS best mentor award goes tooooo.. @kaisakajala.bsky.social !! I'm a bit biased but I believe it was really well deserved!!! 🤣
10.04.2024 18:41 — 👍 4 🔁 0 💬 0 📌 1Thanks!! I reposted it!
04.03.2024 20:48 — 👍 1 🔁 0 💬 1 📌 0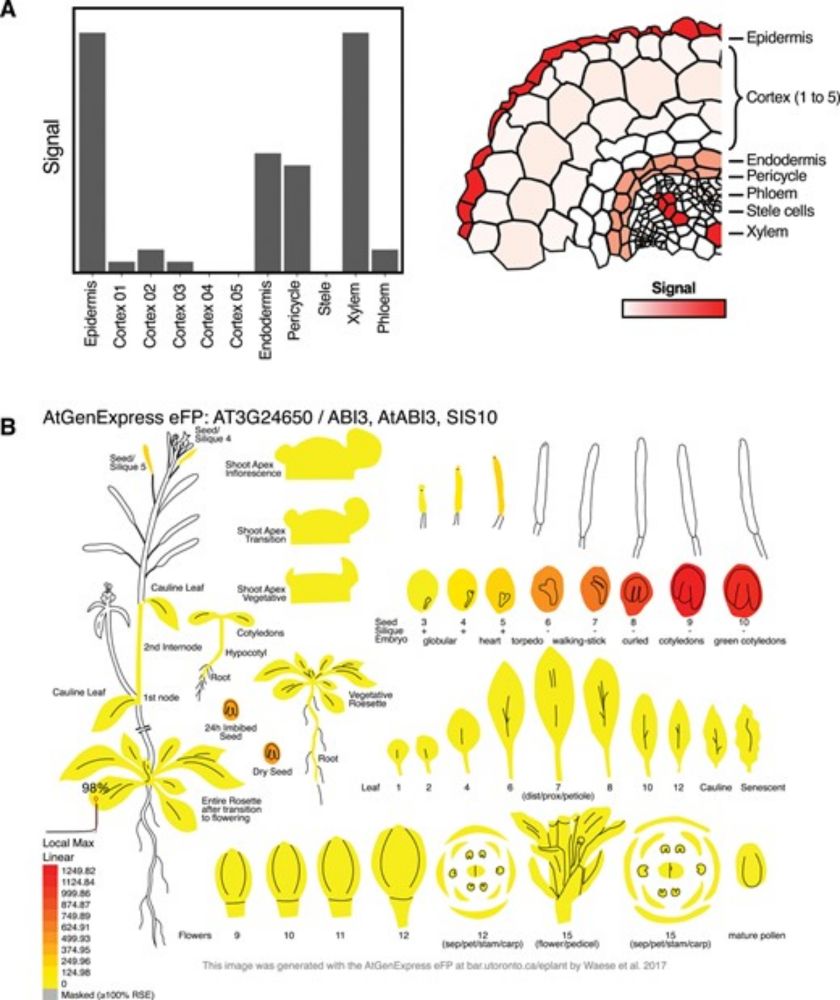
The final version of the ggPlantmap is out at @jxbotany.bsky.social
academic.oup.com/jxb/advance-...
Come for the information, stay for the beautiful figures!
We hope it can encourage people to create their own eFP-viewer!
The final version of the ggPlantmap is out at
@JXBot
academic.oup.com/jxb/advance-... Come for the information, stay for the beautiful figures! We hope it can encourage people to create their own eFP-viewer!


Everything makes sense now...
#chickpea #plantbio
Thanks!!!!
15.02.2024 15:00 — 👍 2 🔁 0 💬 0 📌 0I'll be happy to try something out if you have already a sample file! Let me know if you have something in mind!
All the best!
Hello Javier,
It would be super nice to have an automated tool, but finding one-fit-all segmentation tool would be really challenging. What I wanted to try is to check the output file of different automated tracing tools (microscopic, leaf shape,etc) and check how to convert them into a ggplot map.

ggPlantmap
github.com/leonardojo/g...
Convert Icy-traced Plant images into ggPlot
Integrable with bulk or sc-RNAseq data
Generate .svg for Plant eFP viewer
@jxbotany.bsky.social 2024
academic.oup.com/jxb/advance-...

aRt: Building a root barrier. Built using: ggPlantmap and rayshader #plantbio #bioart
07.02.2024 11:23 — 👍 5 🔁 2 💬 0 📌 0This work was co-authored with Julie Pelletier, and concludes the main works I've done during my PhD in John's lab! We hope that this data is useful for people! Let me know if you have comments or suggestions, I'll be happy to address them.
28.01.2024 14:07 — 👍 0 🔁 0 💬 0 📌 0

GmWRI1 and GmLEC1 target each other in a positive forward loop to promote FA biosynthesis in seeds. We explored a bit the relationship between these two "master" regulators of seed lipid biosynthesis.
28.01.2024 14:07 — 👍 0 🔁 0 💬 1 📌 0
In a transient expression system, we evaluated the function of each motif in GmWRI1 binding sites. To our surprise, mutations in the AW-Box not always resulted in the reduction of enhancer activity, and mutations in the CNC-Box affected the transactivation of CAC2 by GmWRI1.
28.01.2024 14:06 — 👍 0 🔁 0 💬 1 📌 0
To further confirm these results, we identified the list of downregulated genes in the Arabidopsis wri1 mutants and upregulated genes by overexpressing GmWRI1 and identified that indeed, both AW-Box and CNC-Box to be enriched in regions downstream the TSS in Arabidopsis as well.
28.01.2024 14:06 — 👍 0 🔁 0 💬 1 📌 0
Motif analysis revealed the presence and enrichment of the AW-Box in WRI1 binding sites. It has been shown that this element is bound by WRI1 in vitro. Interestingly we found the presence of another element that we called CNC-Box (CNCCNCC).
28.01.2024 14:05 — 👍 0 🔁 0 💬 1 📌 0
When looking at the binding pattern of WRI1, we found that this TF binds preferentially at regions downstream the TSS. This pattern is different from other TFs we studied in the past (LEC1). What could be mechanistic and evolution relevance of this binding pattern?
28.01.2024 14:05 — 👍 0 🔁 0 💬 1 📌 0
After many failed attempts, we finally obtained a good GmWRI1 antibody that allowed us to perform a ChIP-Seq that passed ENCODE QC. With this dataset identified the list of potential target genes of GmWRI1 As expected, these genes encoded for many lipid related genes in seeds.
28.01.2024 14:04 — 👍 0 🔁 0 💬 1 📌 0It is a pretty descriptive work, but we hope to provide resources and insights that can help people who are studying ways to improve oil content in seeds. We will make all the datasets (ChIP-seq, RNA-seq) available upon final publication.
28.01.2024 14:03 — 👍 0 🔁 0 💬 1 📌 0I’m happy to finally submit the final chapter of my PhD dissertation. Here we studied a very known TF in the field of seed lipid content - WRINKLED1. To our surprise, this was the first comprehensive study of genome-wide binding pattern of WRI1. #plantbio #seeds #lipids
28.01.2024 14:01 — 👍 4 🔁 2 💬 1 📌 0I would love to see ggPlantmaps created by the community being uploaded into biocons (bioicons.com) or plant illustrations (figshare.com/authors/Plan...) Can't wait to see! #ggPlantmap #bioicons #dataviz
17.01.2024 22:16 — 👍 0 🔁 0 💬 0 📌 0
Note: ggPlantmap small update (v1.1).
ggPlantmap.to.SVG() converts your ggPlantmap into a SVG file that is compatible with an ePlant EFP pipeline or any graphic software (like powerpoint). The polygons will be grouped based on an assigned column.
Our latest preprint: postdoc Pierre Gautrat devoted his EMBO fellowship to resolving how nutrient and light cues interact to control shade avoidance. With @andruromanowski.bsky.social. The answer is PIF-dependent regulation of cytokinin response regulators. Read all about it in this preprint.
23.12.2023 09:31 — 👍 11 🔁 5 💬 0 📌 1