Does the noncoding genome actually carry more genetic information than coding seqs? Motivated by this question we mutated every bp in the 10kb MYC locus. Results are even more exciting: Decoding the MYC locus reveals a druggable ultraconserved RNA element www.biorxiv.org/content/10.6...
31.01.2026 01:13 — 👍 125 🔁 47 💬 4 📌 6
Huge thanks to @maxewilkinson.bsky.social for making this work possible! (4/4)
23.01.2026 16:17 — 👍 2 🔁 0 💬 0 📌 0

In the process of solving this mystery, we discovered a new, atypical conformation of the spliceosome and discovered that the spliceosome has a surprising propensity to reassemble on excised linear introns in both stressed and unstressed conditions. (3/4)
23.01.2026 16:17 — 👍 2 🔁 0 💬 1 📌 0
In budding yeast cultured under saturation or other prolonged stresses, ~10% of introns accumulate post-splicing as stable, linear RNAs that are protected by the spliceosome. We set out to understand how these stable introns remain associated with the spliceosome and escape canonical RNA decay.(2/4)
23.01.2026 16:14 — 👍 0 🔁 0 💬 1 📌 0
We’re excited to share our latest preprint on the mechanism of excised linear intron stabilization in yeast! This work was led by PhD student @glennli.bsky.social and was a wonderful collaboration with @maxewilkinson.bsky.social. Link: www.biorxiv.org/content/10.6... (1/4)
23.01.2026 16:14 — 👍 58 🔁 25 💬 1 📌 2
First preprint of the year! New work from @jimmy-ly.bsky.social revealing unexpected roles for 5' UTR length in controlling alternate translational isoforms - important implications for both physiological cell function and rare disease. Small changes -> big impacts.
www.biorxiv.org/content/10.6...
23.01.2026 11:39 — 👍 40 🔁 13 💬 0 📌 0
We are looking for a postdoc to work on mechanisms of #RNA decay in cancer using #cryoEM with #nanobodies and #minibinders! Please RT
18.01.2026 14:30 — 👍 22 🔁 27 💬 2 📌 0
We find that disrupting the miR-200–ZEB1 double-negative feedback loop leads to anovulatory infertility and widespread gene expression changes in the mouse pituitary. This study demonstrates the dramatic phenotypic and molecular consequences of disrupting repression of a single miRNA target. (2/2)
14.01.2026 21:11 — 👍 3 🔁 0 💬 0 📌 0

Check our latest collaboration with the Kleaveland Lab (kleavelandlab.org), led by Joanna Stefano and Lara Elcavage: academic.oup.com/nar/article/... (1/2)
14.01.2026 21:11 — 👍 14 🔁 4 💬 1 📌 1
Huge thanks to Brenda Schulman and @jakobfarnung.bsky.social for the exceptionally collaborative effort from start to finish. We also thank @wyppeter.bsky.social, Lianne Blodgett, and Daniel Lin for their invaluable contributions to this work! (5/5)
06.01.2026 15:16 — 👍 3 🔁 0 💬 0 📌 0
Our results establish AGO binding and polyubiquitylation as the key regulatory steps of TDMD, define a unique class of cullin–RING E3 ligases that depend on CUL3 and ELOB/C, and reveal generalizable RNA- and protein-mediated interactions that specify AGO degradation with exquisite selectivity. (4/5)
06.01.2026 15:16 — 👍 0 🔁 0 💬 1 📌 0
We demonstrate selective binding of ZSWIM8 to a human AGO–microRNA–trigger complex for CUL3-mediated polyubiquitylation of the AGO protein. Furthermore, cryo-EM analyses reveal how ZSWIM8 recognizes the distinct AGO and RNA conformations shaped by pairing of the microRNA to the trigger. (3/5)
06.01.2026 15:16 — 👍 0 🔁 0 💬 1 📌 0
The ZSWIM8 E3 ligase was known to cause degradation of AGO–microRNA complexes bound to trigger RNAs. However, whether and how ZSWIM8 directly recognizes these complexes among the preponderance of non-trigger-bound AGO–microRNA complexes in the cell has been a mystery. (2/5)
06.01.2026 15:16 — 👍 0 🔁 0 💬 1 📌 0
We are thrilled to share our latest work uncovering the mechanistic basis of target-directed microRNA degradation (TDMD). This work was driven by @jakobfarnung.bsky.social and @elenaslo.bsky.social in a fantastic collaboration with Brenda Schulman's lab. tinyurl.com/E3TDMD (1/5)
06.01.2026 15:16 — 👍 89 🔁 29 💬 2 📌 4
We identify 5 sites in 3' UTRs of mRNAs that trigger target-directed microRNA degradation (TDMD) of miR-335-3p, miR-322, and miR-503, uncovering noncoding functions of these mRNAs. This study positions TDMD within imprinted gene networks on the battleground of parental conflict (2/3)
07.11.2025 18:33 — 👍 2 🔁 0 💬 1 📌 0
We've uncovered two mechanisms that coronaviruses use to solve the “tailomere problem” and identified an mRNA degradation pathway that operates independently of viral protein nsp1. Many thanks to Eugene Valkov's lab (@eugenevalkov.bsky.social) for their help with this study.
13.10.2025 19:44 — 👍 4 🔁 0 💬 0 📌 0
Since their discovery, we have known lysosomes possess RNase activity; however, their endogenous substrates were not known. Surprisingly we found preferential targeting of specific RNAs for lysosomal degradation by autophagy and identified sequence motifs that mediate their lysosomal targeting (2/2)
11.09.2025 13:59 — 👍 3 🔁 0 💬 0 📌 0
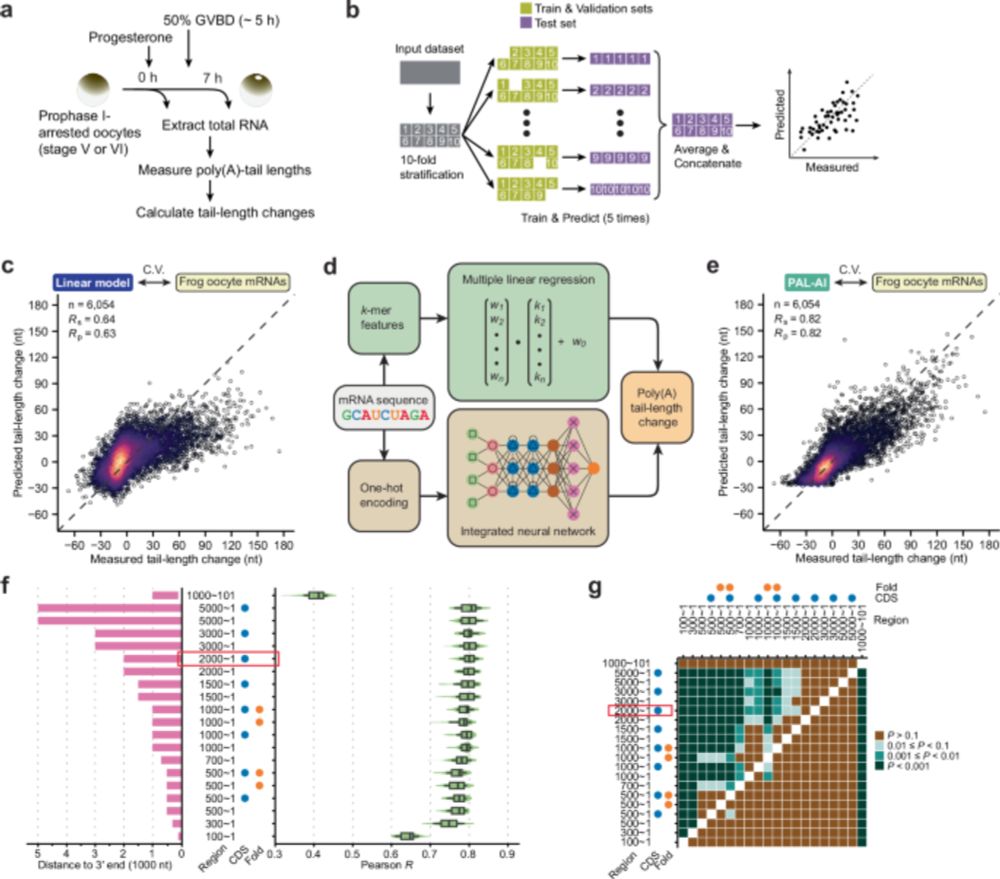
We developed a neural network machine-learning model that predicts poly(A) tail-length changes in frog, mouse, and human oocytes, revealing new regulatory motifs and showing that variants disrupting tail lengthening are under negative selection, thus linking tail-length control to human fertility.
02.08.2025 15:01 — 👍 0 🔁 0 💬 0 📌 0
Don’t miss this Q&A with Dr. Michelle Frank (@michelle-frank.bsky.social), an awesome postdoc in our lab!
05.04.2025 19:51 — 👍 18 🔁 2 💬 0 📌 0
We report that some miRNAs are capable of high affinity binding to 3′-only sites (stretches of extensive perfect pairing to the 3′ region, without any pairing to the miRNA seed) and that these sites are functional, imparting post-transcriptional repression to site-containing reporter mRNAs. (2/2)
15.03.2025 00:40 — 👍 6 🔁 0 💬 0 📌 0
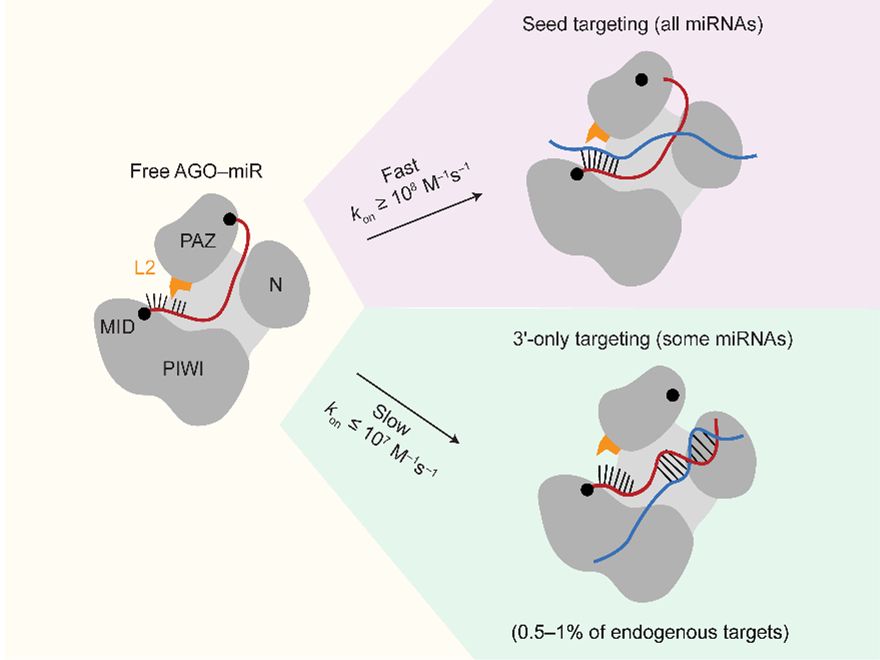
Check out the latest study from our lab, led by @mhall98.bsky.social :
www.biorxiv.org/content/10.1... (1/2)
15.03.2025 00:36 — 👍 48 🔁 17 💬 1 📌 1
Happy 2025! Excited to finally share our published slicing structure of human AGO2, the catalytic structure for RNAi by siRNAs and miRNAs. This was an amazing collab effort between @voslab.org and @bartellab.bsky.social with @amohamed98.bsky.social
01.01.2025 18:31 — 👍 9 🔁 2 💬 2 📌 0
keen on reverse transcription | also keen on spliceosomes | cryoEM dabbler
Assistant Member @MSKCC
Formerly post-doc @MIT
Formerly-formerly PhD @MRC_LMB
Formerly formerly-formerly Otago Uni
Kiwi 🥝 🇳🇿
https://wilkinsonlab.bio/
Assistant Professor at RNA Therapeutics Institute, UMass Chan Medical School
Assistant Professor at University of Central Florida. Stressing bacteria, pesting pathogens and folding RNA. Foodie, music lover and traveller. Fier Québécois.
PostDoc @Doudna_lab 🧬| PhD @Cambridge_Uni | @Harvard @imperialcollege - I like epigenetics and editing genes. Life begins at the end of your comfort zone✨✈️🌍⛵️🎿
Transcription and RNA processing; postdoc in @stoplab.bsky.social group @uam-ibmib.bsky.social
ASBMB drives discovery forward in academic, industry and governmental sectors, helping to generate new scientific knowledge today that fuels tomorrow’s biotechnological and medical breakthroughs.
www.asbmb.org | #ASBMB26
Scientist in Dept of Mol Bio, UTSW/HHMI, studying post-transcriptional regulation & noncoding RNAs. Opinions are my own and do not reflect those of my employer.
https://labs.utsouthwestern.edu/mendell-lab
https://www.hhmi.org/scientists/joshua-t-mendell
PhDing in the @CorcoranLab, dabbles with RNA granules, viruses, and the outdoors
Fosters productive collaborations, rigorous debate, and enduring friendships between RNA scientists across Colorado. Fueled by 🍕, 🍺, and 🧬 since 1986.
PhD candidate @HarvardChemBio in the Flynn Lab @BCHStemCell 🧬🍬👩🔬
RNA-curious PhD student | Mühlemann Lab | NCCR_RNADisease
I’m just a girl in an RNA world
[alternative splicing]
MD-PhD student @jacksonlab.bsky.social, @uconnresearch.bsky.social
JCC fellow and Postdoc in the McLellan Lab at UT Austin
Postdoc in the Santos Lab (The Francis Crick Institute)
Investigating how cells navigate change
Trying to appreciate the beauty in nature 🌱
Postdoc Jonathan Weissman lab | PhD Feng Zhang lab
We study RNA Biology using a variety of approaches.
medicine.yale.edu/lab/steitz/