
This week in the HUJI PES seminar:
@hebrewuniversity.bsky.social
@finkel-lab.bsky.social
Microbial ecology and plant-microbe interactions at @HebrewU http://finkel-lab.com

This week in the HUJI PES seminar:
@hebrewuniversity.bsky.social

🎉 Check out the latest from Qin Gu’s team at Nanjing Agricultural University: “Keystone Pseudomonas species in the wheat phyllosphere” (Cell Host & Microbe) 👉 www.cell.com/cell-host-mi...
23.11.2025 08:11 — 👍 13 🔁 10 💬 1 📌 0Wish I could’ve been there for that :)
13.11.2025 14:51 — 👍 1 🔁 0 💬 1 📌 0I remember Aharon destroying this paper in an epic departmental seminar just a couple weeks after it was published.
13.11.2025 07:40 — 👍 1 🔁 0 💬 2 📌 0
This week in the HUJI PES seminar:
@hebrewuniversity.bsky.social
I'm thrilled to share our newest publication in @natmicrobiol.nature.com, led by Drs. @botanichole.bsky.social, Valéria Custódio, and David Gopaulchan: Precipitation legacy effects on #SoilMicrobiota facilitate adaptive drought responses in plants. 🌾 🧪 (thread)
www.nature.com/articles/s41...

Call for Applications for the Azrieli International Postdoctoral Fellowship-2026-27 azrielifoundation.org/azrieli-fell...
09.11.2025 08:29 — 👍 4 🔁 4 💬 1 📌 0
The first time we are hearing about viruses at #PMS2025, thanks @sheilaroitman.bsky.social!
07.11.2025 10:55 — 👍 9 🔁 2 💬 2 📌 0Fascinating talk on how to optimize microbe-microbe interactions with Bacillus as the focal species.
06.11.2025 14:24 — 👍 2 🔁 0 💬 0 📌 0
POV running with Asaf
05.11.2025 11:18 — 👍 3 🔁 0 💬 0 📌 0I approve this message :)
04.11.2025 15:35 — 👍 2 🔁 0 💬 0 📌 0
Check out our latest preprint in which we describe a new phage plaque sequencing pipeline which lowers the cost of sequencing by up to 10-fold while significantly shortening the time required to obtain the phage genome sequences
www.biorxiv.org/content/10.1...

This week in the HUJI PES seminar:
@hebrewuniversity.bsky.social

This week in the HUJI PES seminar:
@hebrewuniversity.bsky.social
In plant-pathogen or plant-microbe interactions what is the proper null.
- there is specific coevolution of any pathogen/microbe such that it was at least partly adapted to the host isolated.
-the host of isolation may have not effected the pathogen in the slightest.
New pre-print from the team!
The manuscript is @emma-raven.bsky.social's PhD work showing that whether a leaf is a carbon sink or a carbon source influences how they execute immune responses.
Have a read!
#PlantScience
@johninnescentre.bsky.social

The team at our research site in the alps
📣 New PhD student position opening in my lab!
Soils are full of microbes, but how many are active?
Surprisingly, we still lack reliable methods to answer this.
If you are interested in microbial dormancy and are fascinated by the Alps and glacier forefields, contact me.
🏔️ 🦠 💤 🧬 🎓

Our paper, describing how the T6SSs of P. putida shape the tomato rhizosphere, is now in its final format in @isme-microbes.bsky.social ISME Communications. If you'd like to learn more, here is a thread (1/11) or read the Article doi.org/10.1093/isme....
06.10.2025 18:22 — 👍 55 🔁 19 💬 2 📌 0
Original article: www.nature.com/articles/s41...
08.10.2025 17:10 — 👍 0 🔁 0 💬 0 📌 0
Check out this preview @naturalist1986.bsky.social and I wrote about the fantastic paper on wood microbiota from Wyatt Arnold and colleagues: www.sciencedirect.com/science/arti...
08.10.2025 17:10 — 👍 4 🔁 4 💬 1 📌 0
Time-series metatranscriptomics reveals differential salinity effects on the methanogenic food web in paddy soil | mSystems journals.asm.org/doi/full/10....
23.07.2025 05:09 — 👍 6 🔁 4 💬 0 📌 0
Unannotated translation products are widespread in model E. coli
www.biorxiv.org/content/10.1...

We've tried to take a different view at the issue of nutrient limitations-looking for the shared basis rather than for specific response. This is the result:
www.jbc.org/article/S002...
Led by Hagit Zer with Stav Chen and David Rasin, in close collaboration with Miguel Hernandez–Prieto. Thanks!
Very nice writeup on someone I've admired and learned from for many years. I didn't know he had passed until catching up on This Week in Microbiology driving to campus this morning. His 1958 paper on Salmonella growth rates basically founded the formal study of microbial physiology, which...
12.09.2025 13:46 — 👍 12 🔁 5 💬 1 📌 0So excited to share this new paper by @sulheim.bsky.social and others! It's rare that you find a pattern, propose a hypothesis and the more you look, the more data you find that fits! Snorre's intuition, hard work and rigour to put it all together have been inspiring! Let us know what you think...
22.08.2025 14:05 — 👍 49 🔁 12 💬 1 📌 1
Out after peer review, collaborative study from Nordborg & Weigel labs with help from many others. Not the largest collection of new Arabidopsis thaliana genomes, but we hopefully put forward some good ideas for how to think about pangenomes and their analysis!
www.nature.com/articles/s41...
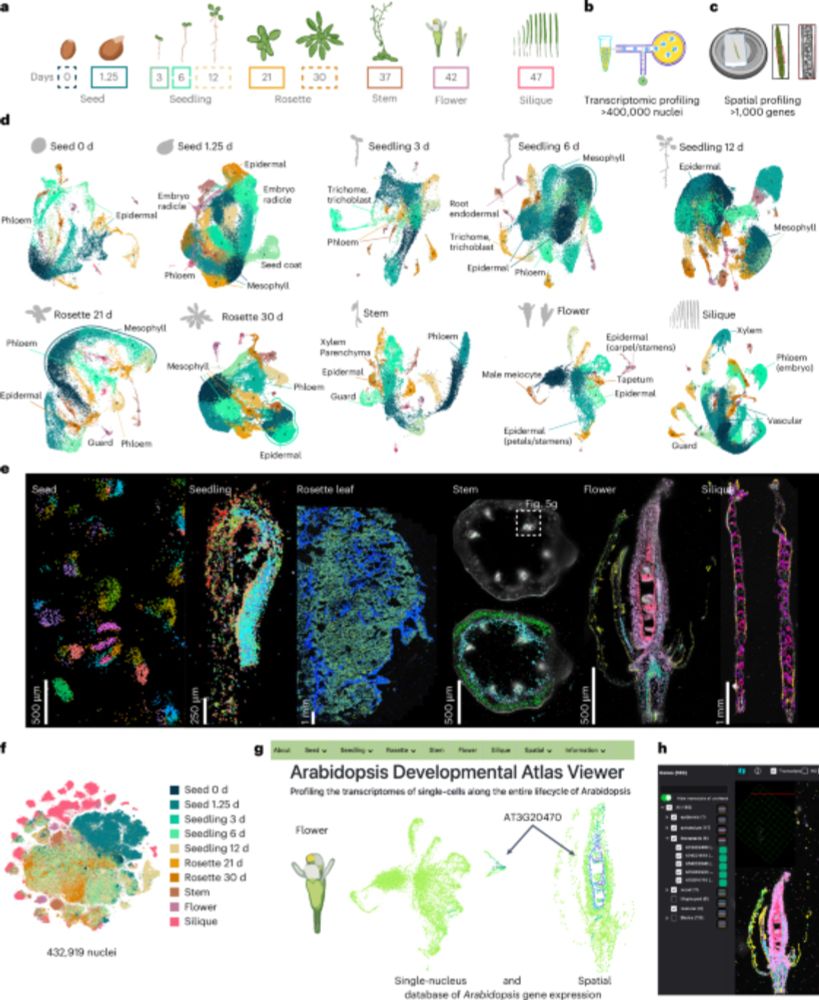
From a gene’s single cell expression–through spatial localization–to novel function, and beyond!
Now out @natplants.nature.com
We built a comprehensive spatial-transcriptomic atlas of Arabidopsis, revealing cell-type identities across organs in unprecedented detail
www.nature.com/articles/s41...

Why chocolate tastes so good: microbes that fine-tune its flavour www.nature.com/articles/d41...
20.08.2025 03:49 — 👍 5 🔁 2 💬 0 📌 0
In memory of Fabrice Rappaport. A myovirus encoding both photosystem I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells www.nature.com/articles/s41...
06.08.2025 11:02 — 👍 6 🔁 2 💬 0 📌 0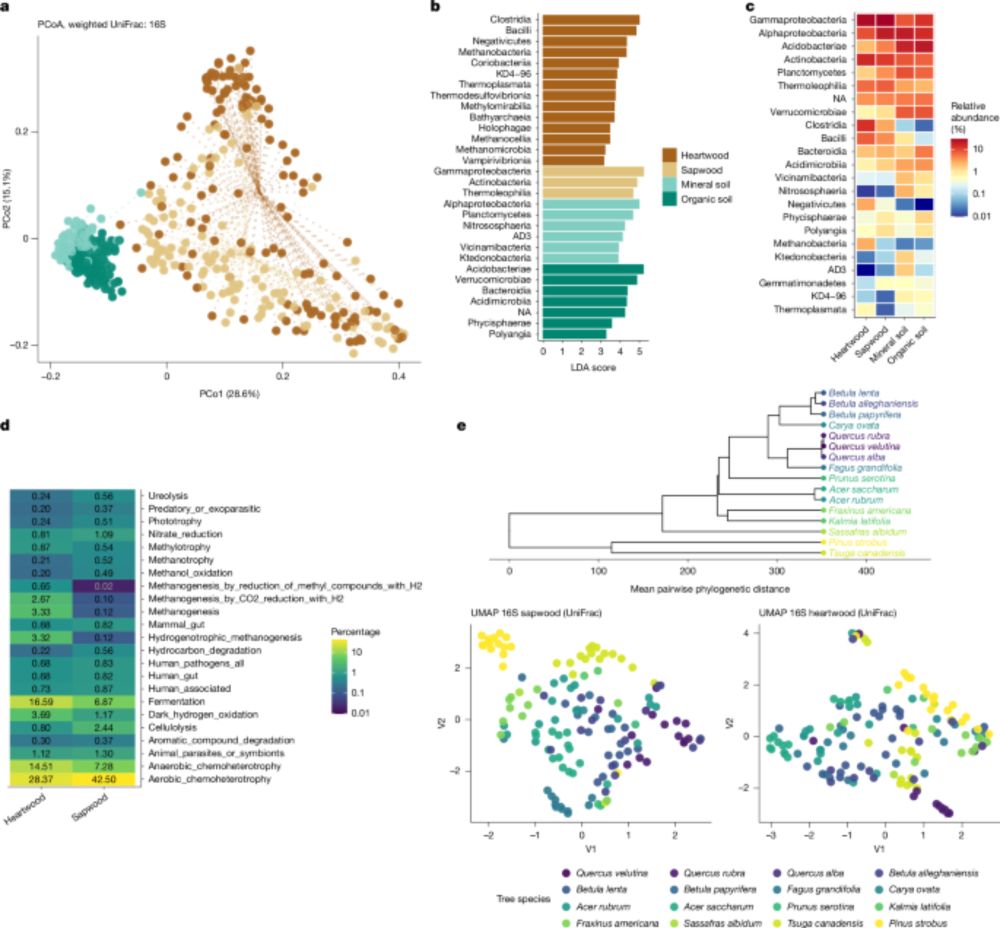
#NatMicroPicks
Hidden microbial world in trees🌳
Living wood hosts trillions of bacteria making trees a complex ecosystems with major roles in forest health and function.
#PlantMicro #MicroSky
www.nature.com/articles/s41...