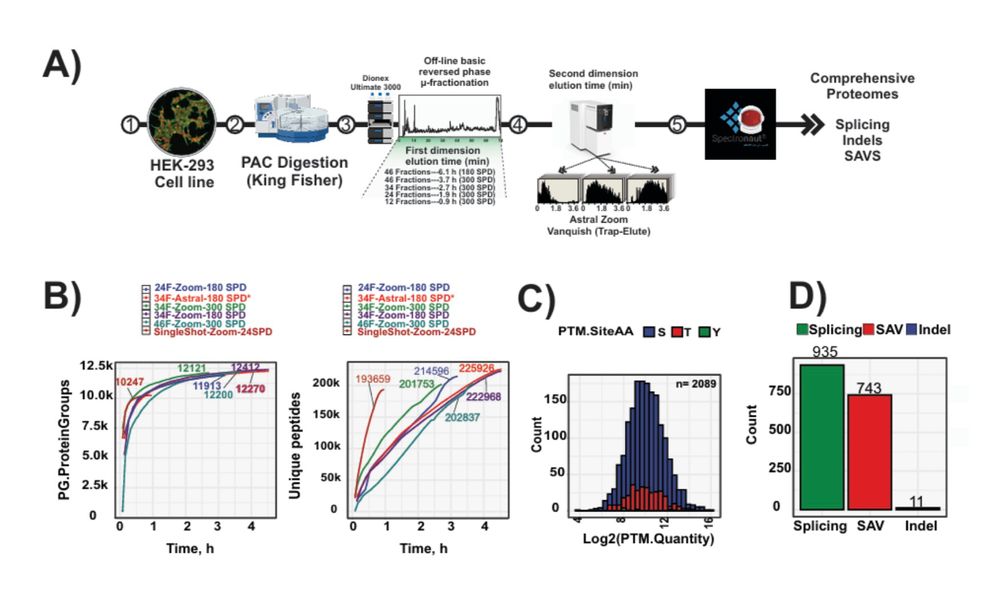
With a 40% reduction in analysis time and >12,000 proteins covered in 2.7 hours, it's setting new benchmarks for proteome studies.
19.07.2025 09:21 — 👍 5 🔁 0 💬 0 📌 0@jesperolsenlab.bsky.social
Mass spectrometry for quantitative proteomics. The Olsen group at the Center for Protein Research, University of Copenhagen.

With a 40% reduction in analysis time and >12,000 proteins covered in 2.7 hours, it's setting new benchmarks for proteome studies.
19.07.2025 09:21 — 👍 5 🔁 0 💬 0 📌 0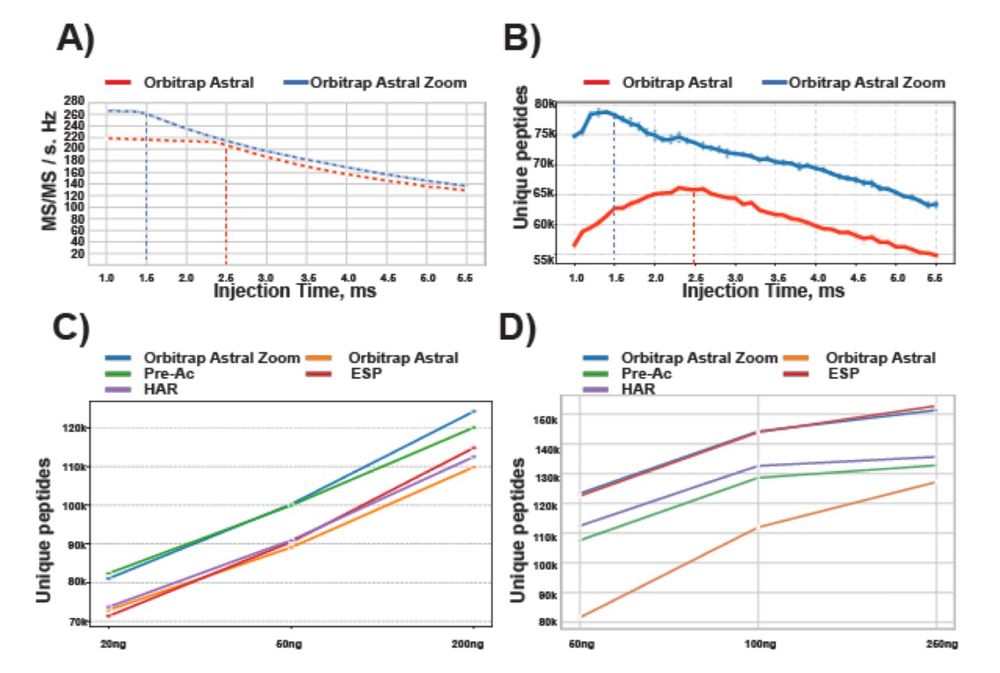
The new Astral Zoom MS overcomes previous MS limitations and is now achieving ultra-fast scans up to 270 Hz. It allows to identity > 100,000 peptides with short LC gradients.
19.07.2025 09:21 — 👍 4 🔁 0 💬 1 📌 0
🚀 Check out our preprint on the new Thermo Scientific Orbitrap Astral Zoom MS 💫👇🏼
www.biorxiv.org/content/10.1...
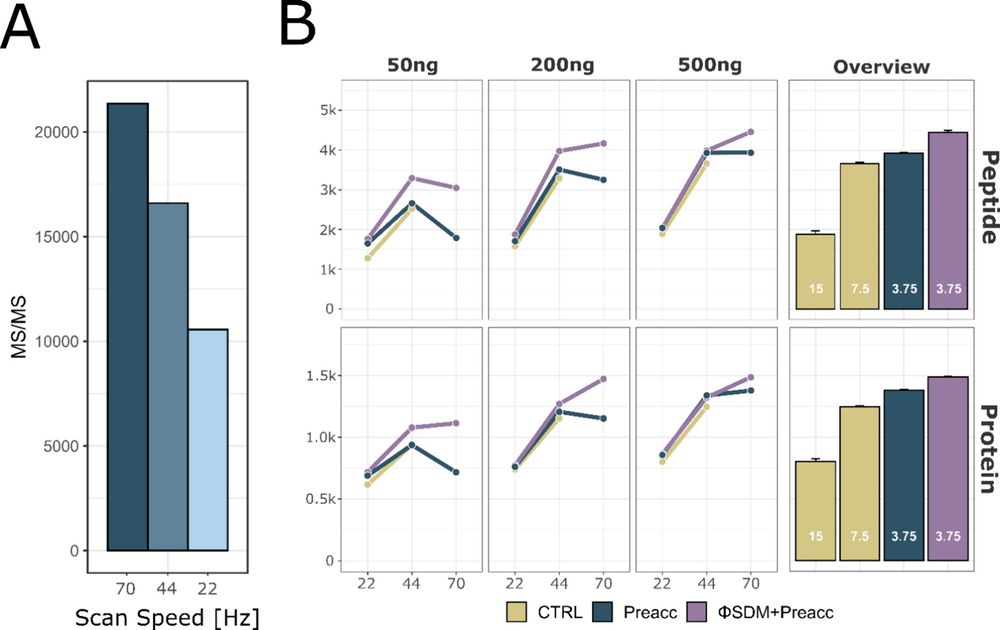
The combination of pre accumulation and faster scans significantly improves peptide/protein IDs on short LC gradients and boosts sensitivity for high-throughput workflows 📈
The benefits are even higher with ΦSDM, especially in fast, low-resolution Orbitrap runs.
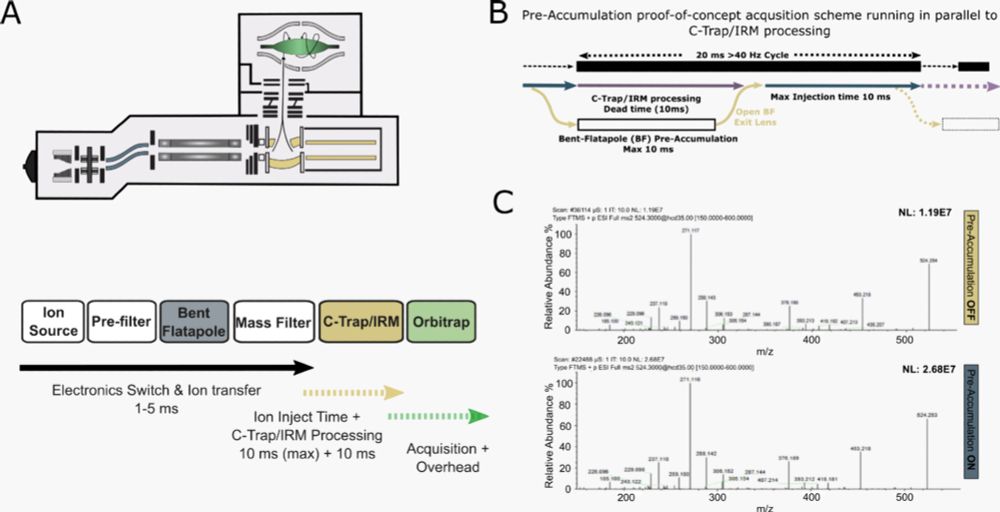
Florian Harking & Ulises H. Guzman made it happen by preaccumulating ions in the bent flatapole and applying phase-constrained spectrum deconvolution method (ΦSDM).
01.07.2025 12:29 — 👍 0 🔁 0 💬 1 📌 0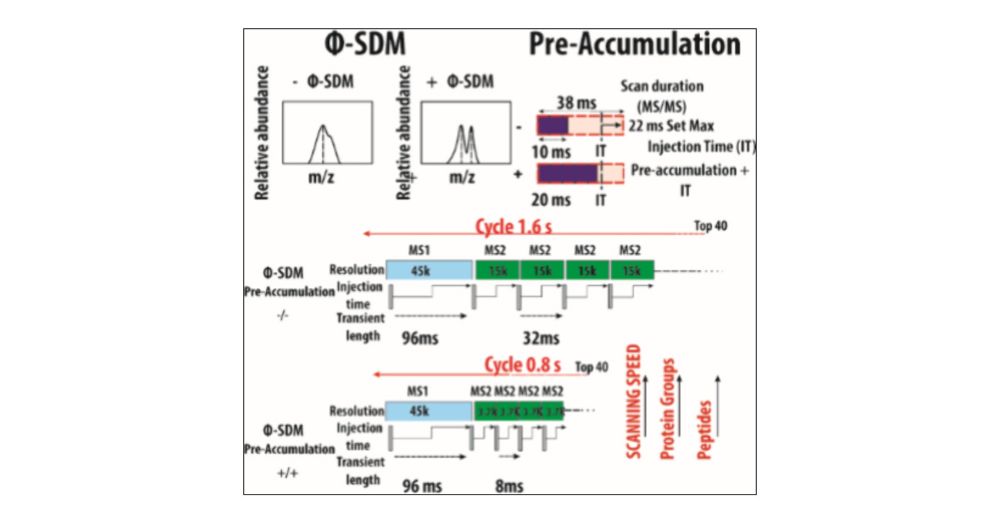
Boosting Orbitrap MS/MS speed to ∼70 Hz? 🚀
Read how we did it in our latest paper 👇
pubs.acs.org/doi/10.1021/...
🦷 Excited to share how proteomics is pushing the boundaries of palaeoanthropology!
We used proteomics on Paranthropus robustus fossils from South Africa (~2 million years old) to get insights into biological sex and variation.
A great example of how LC-MS/MS can uncover new transformative info👇
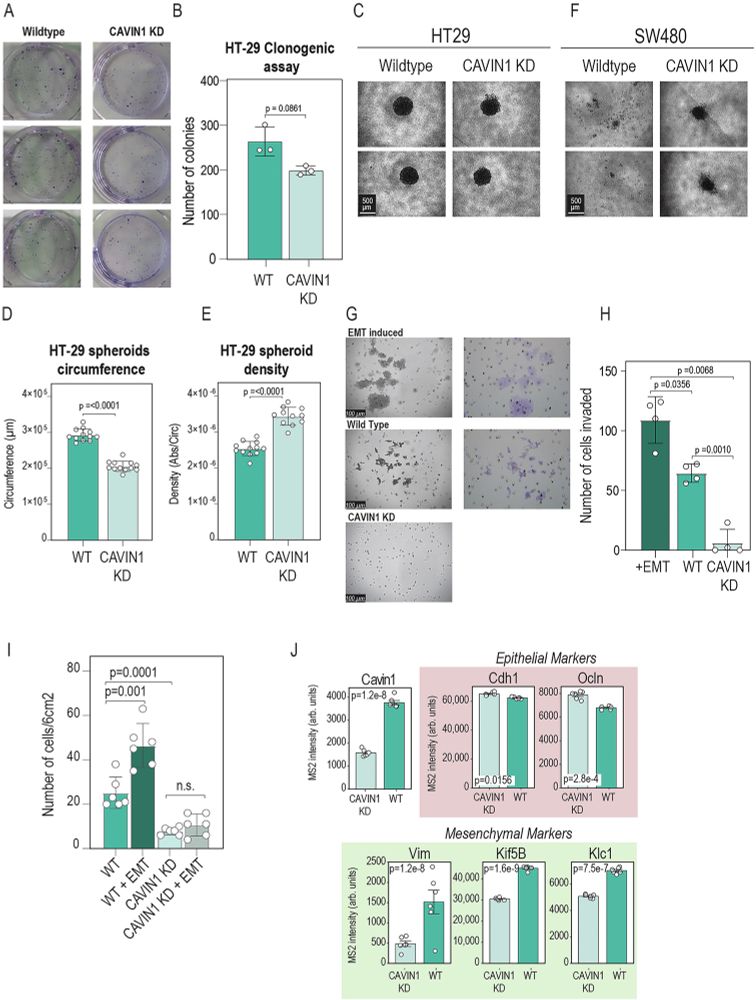
Knocking down CAVIN1 in a 3D model reveals its connection with tumour invasiveness. Hybrid-DIA based phosphoproteomics profiling reveals an intriguing link between mTOR-signalling and its potential association to CAVIN1.
25.04.2025 09:45 — 👍 1 🔁 0 💬 0 📌 0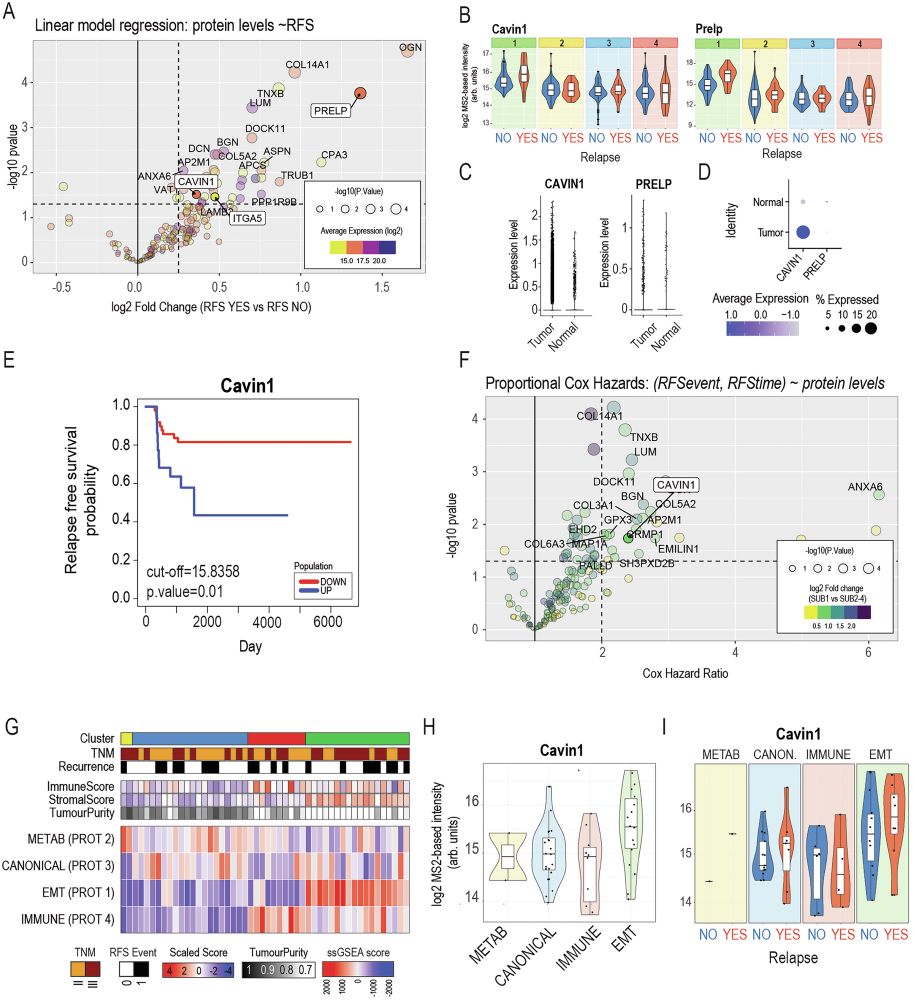
The mesenchymal-like subtype was shown to be marked by increased CAVIN1 and PRELP. Higher CAVIN1 correlates with higher relapse risk in two independent cohorts. We confirmed this in a second cohort and were able to link this to Epithelial to Mesenchymal Transition.
25.04.2025 09:45 — 👍 1 🔁 0 💬 2 📌 0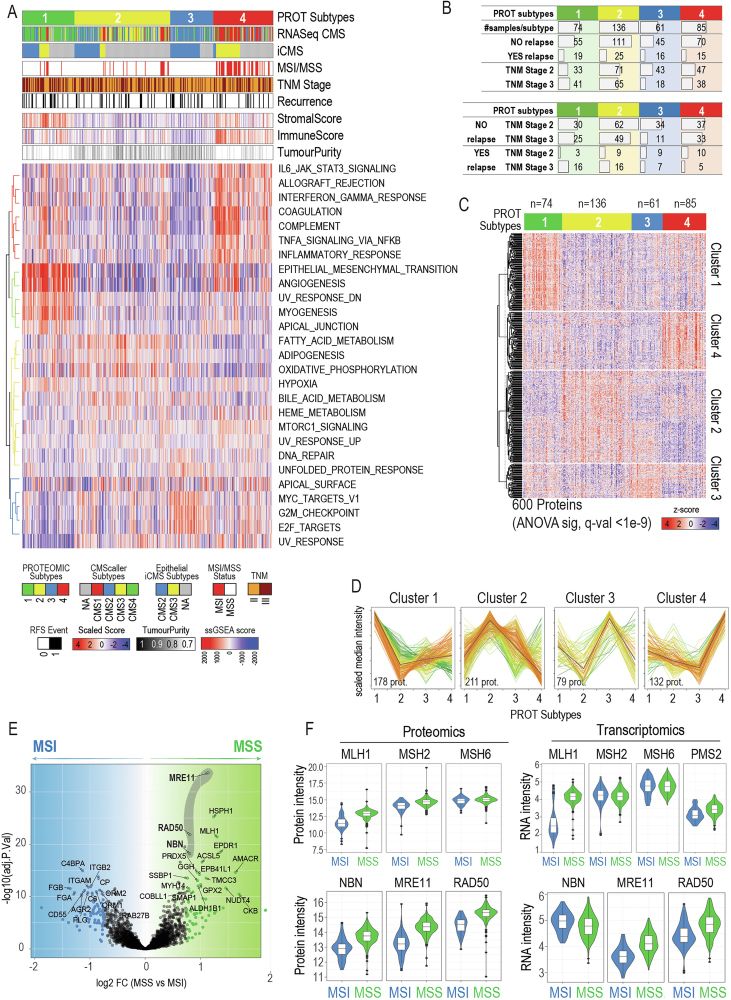
We stratified 412 CRC tumors into 4 distinct subtypes using quantitative proteomics. Our results show clear links to transcriptomics CMS, immune infiltration, tumor purity, and relapse.
25.04.2025 09:45 — 👍 0 🔁 0 💬 1 📌 0
🚨 Check out our new study on proteomic profiling in premetastatic colorectal cancer! Accross 412 tumors, we define molecular subtypes with distinct functional landscapes and revealed CAVIN1 as novel biomarkers for relapse risk.
www.embopress.org/doi/full/10....
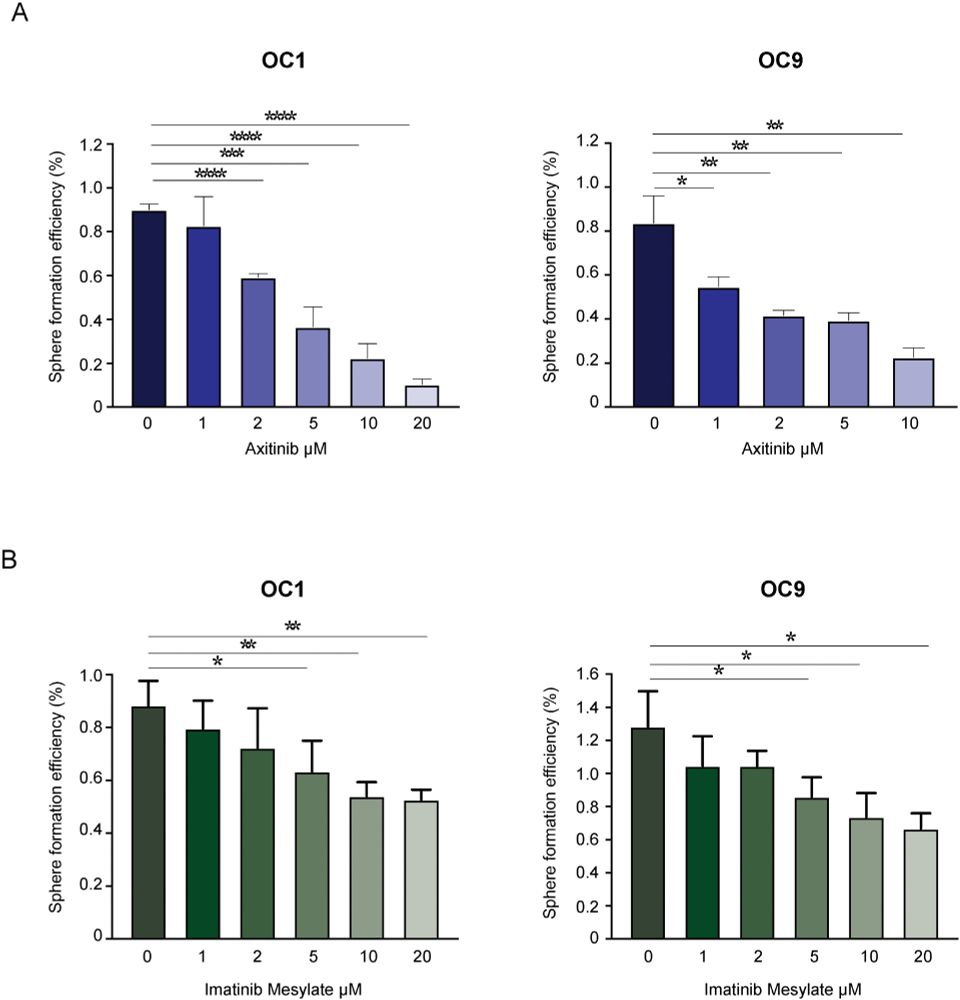
We also identified aberrant PDGF receptor (PDGFR) activation as a key feature of OCSCs.
When we treated patient-derived ovarian cancer cells with a PDGFR inhibitor, their stemness potential dropped (sphere formation ↓).
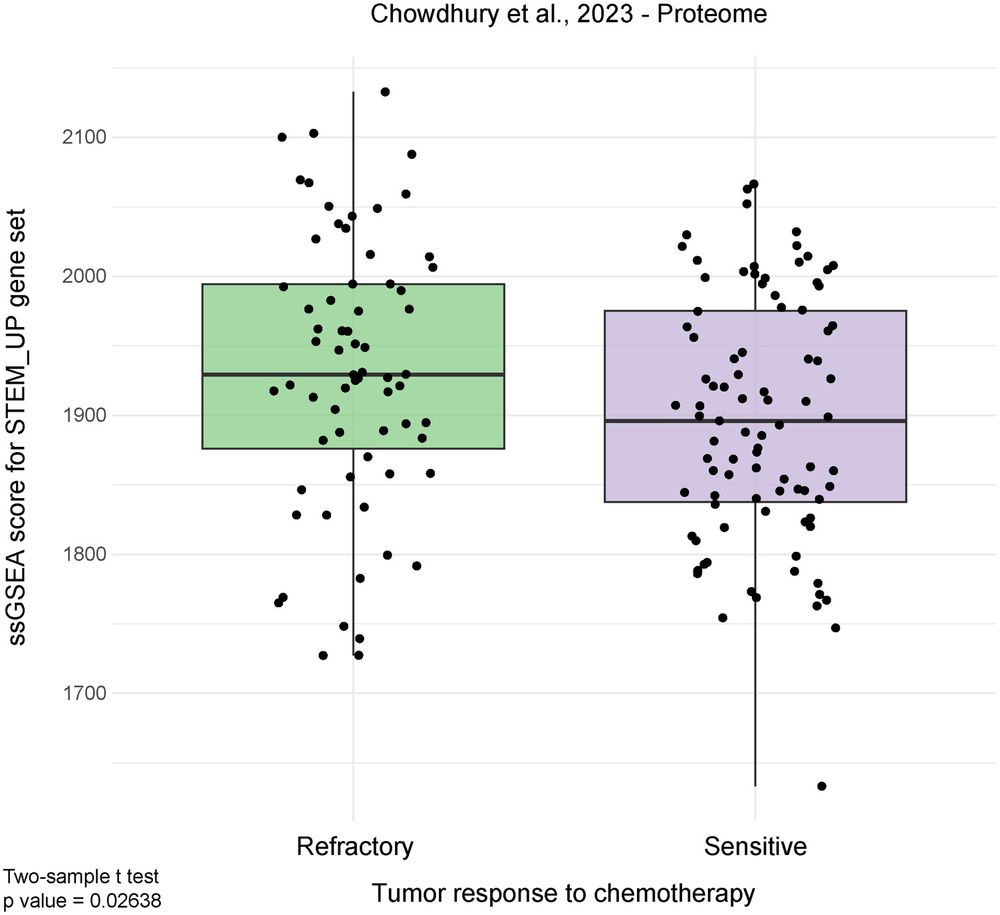
Our stemness signature correlates with chemo-refractoriness in clinical proteomics data.
That’s huge: it means this signature might help predict therapy resistance.
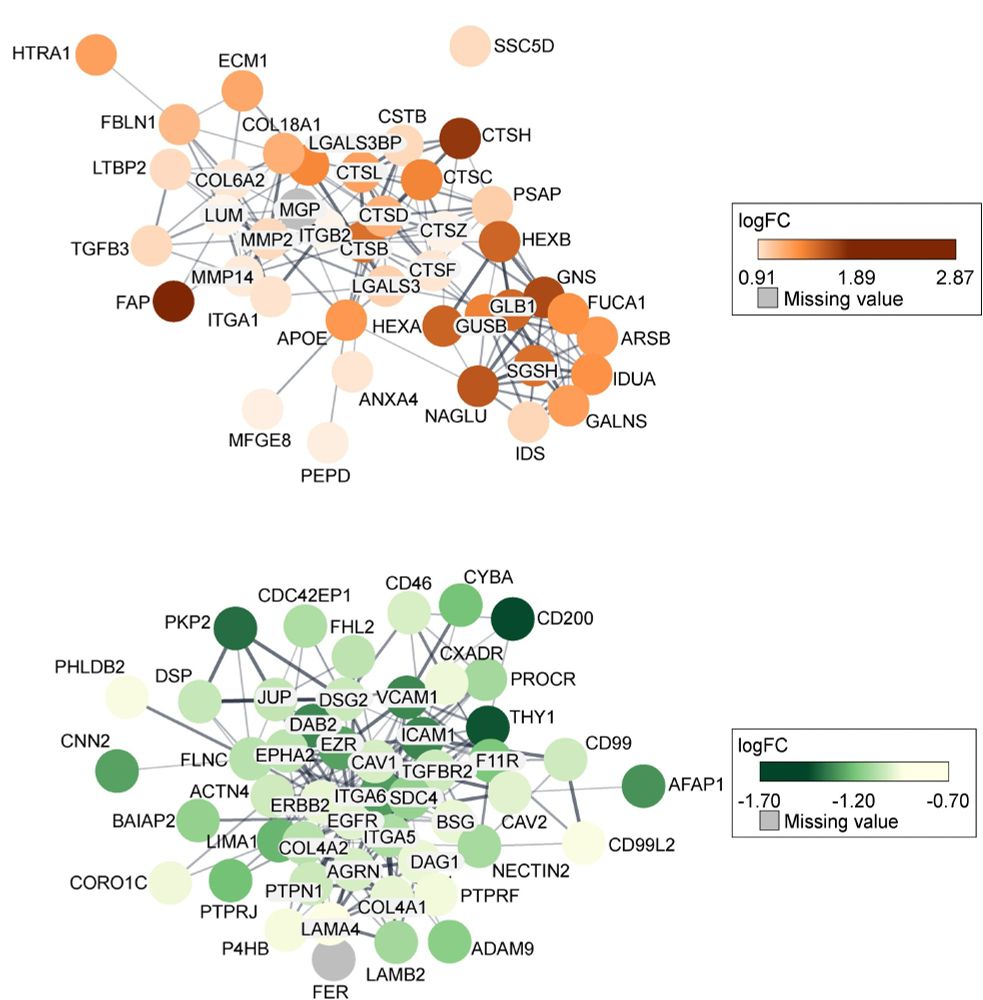
Stemness-associated proteins were involved in extracellular matrix remodeling.
This points to a link between the stem-like state and the tumor microenvironment.
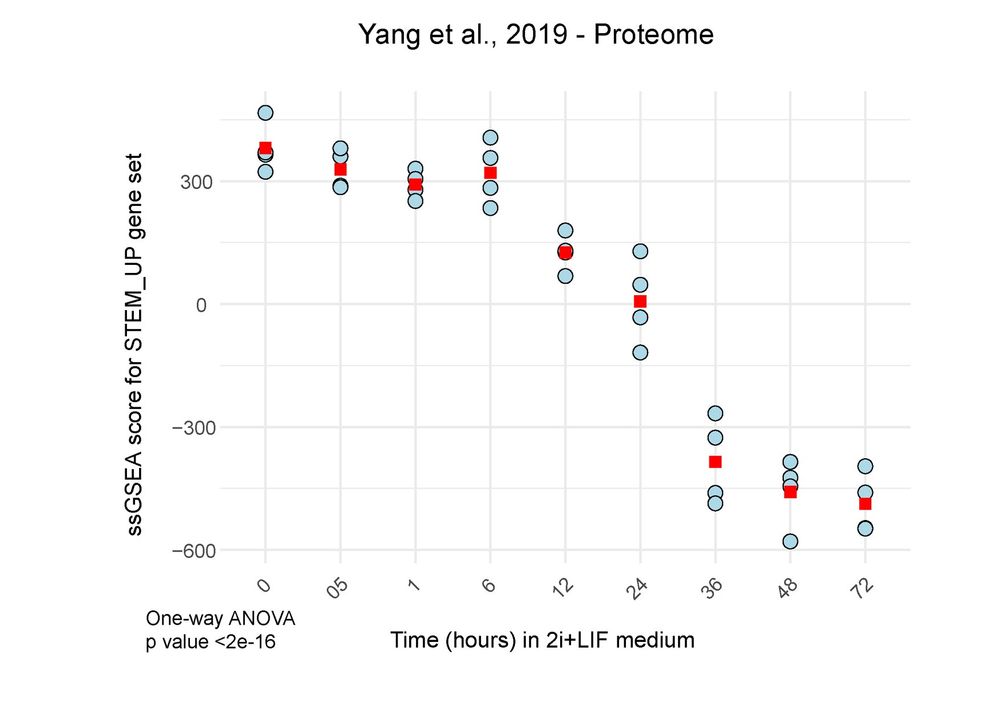
To validate our stemness signature, we used single-sample GSEA on published proteomics data from embryonic stem cell differentiation.
24.04.2025 08:56 — 👍 0 🔁 0 💬 1 📌 0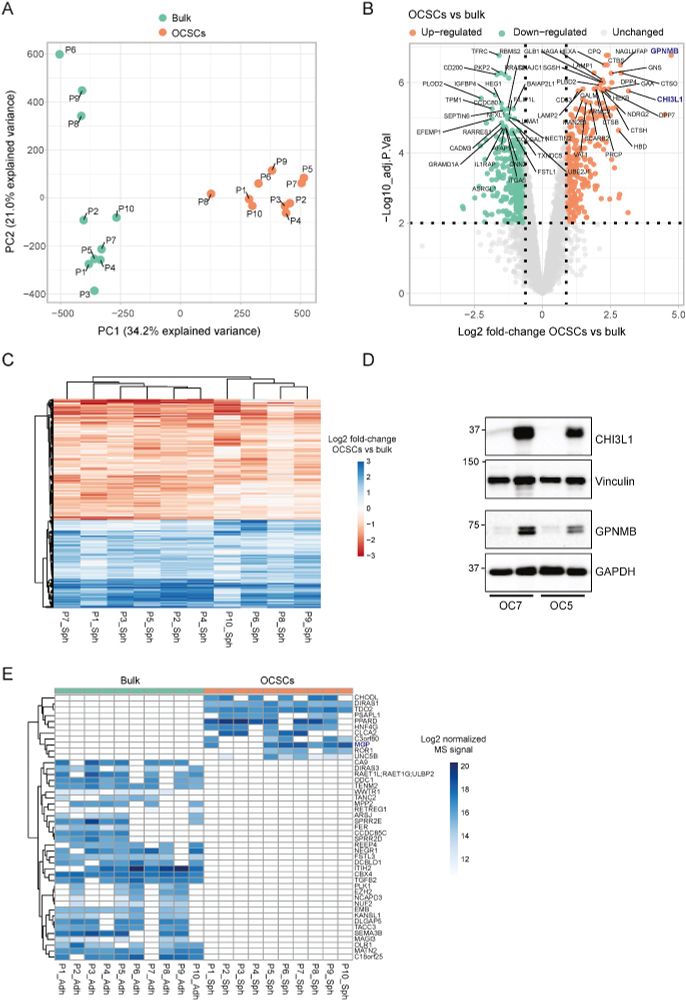
We profiled the proteome and phosphoproteome of 3D spheres enriched for OCSCs from 10 treatment-naive patient-derived lines.
This revealed a shared stemness-related signature across patients, despite high inter-patient heterogeneity.
Our new study is out! 🚨
We used mass spectrometry to uncover a conserved stemness signature in ovarian cancer stem cells (OCSCs).
Check what we found and why it matters. 👇
#OvarianCancer #CancerStemCells #MassSpec #proteomics
www.mcponline.org/article/S153...
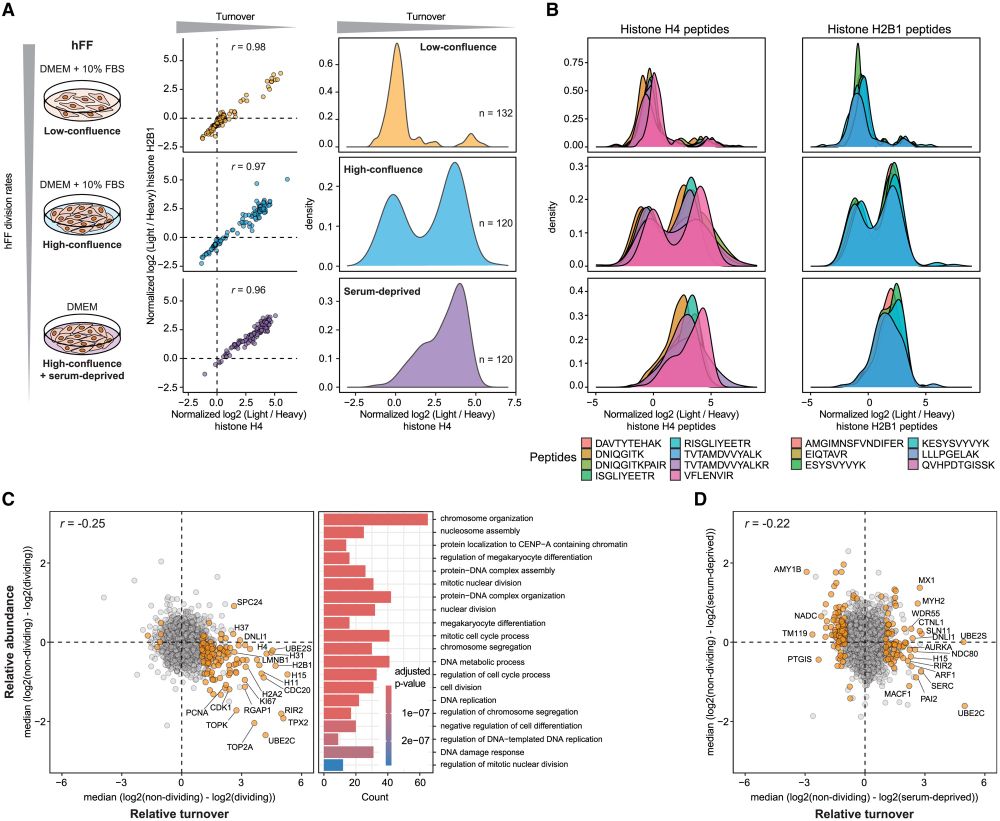
We confirmed this observation and exploited it to compare different modalities of cell division arrest in human foreskin fibroblasts.
31.03.2025 15:11 — 👍 1 🔁 0 💬 0 📌 0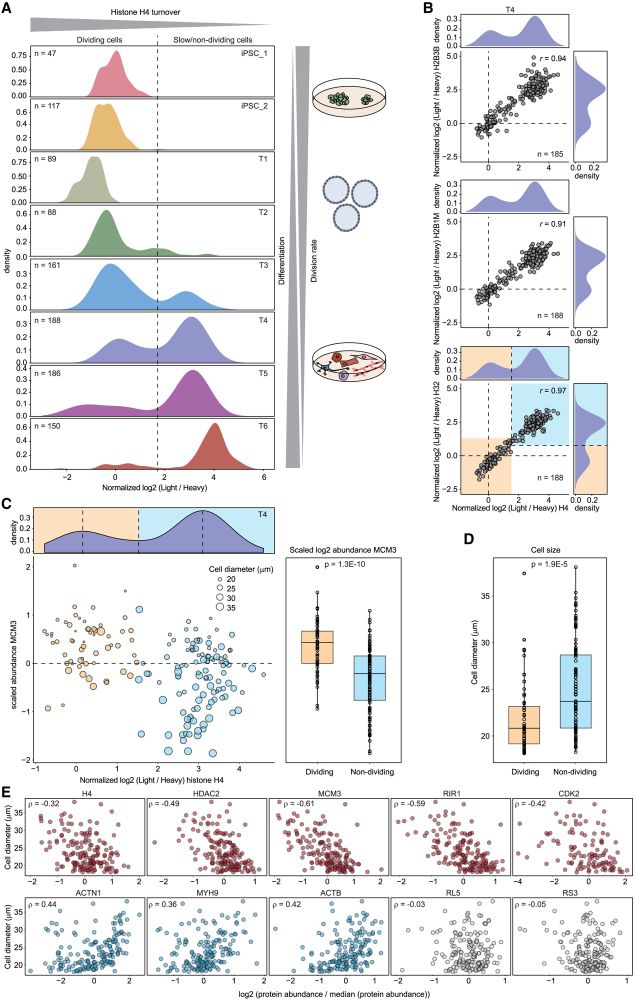
Histone turnover enables distinguishing dividing cells from slow/non-dividing ones, due to the extremely slow core histone turnover in non-dividing cells.
31.03.2025 15:11 — 👍 0 🔁 0 💬 1 📌 0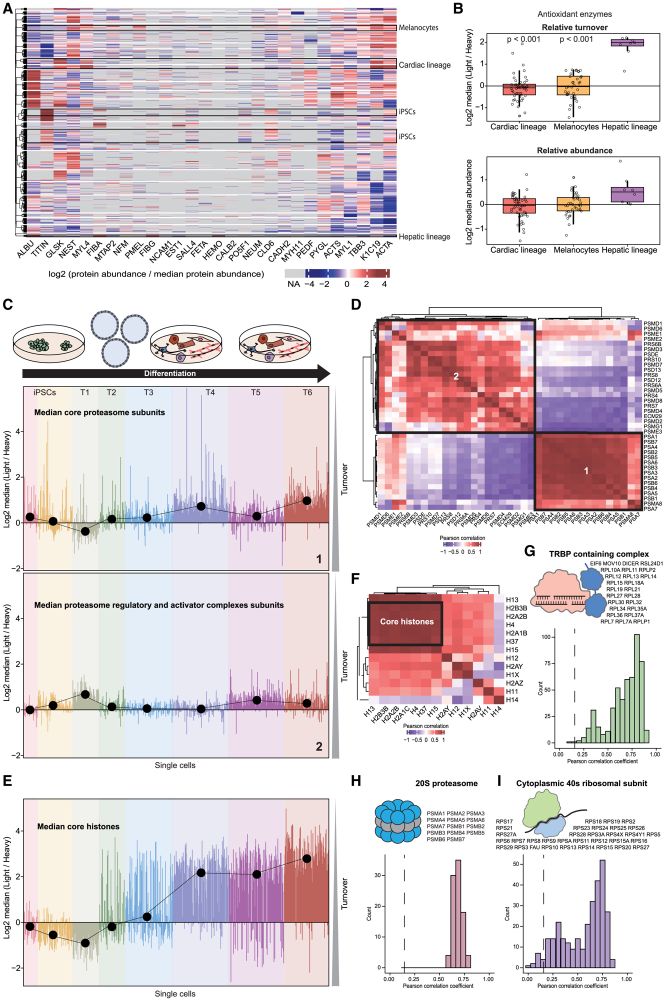
Protein turnover provides an orthogonal measurement to protein abundance, revealing functional differences between cell types and states, and improving protein co-regulation analysis to determine members of protein complexes.
31.03.2025 15:11 — 👍 0 🔁 0 💬 1 📌 0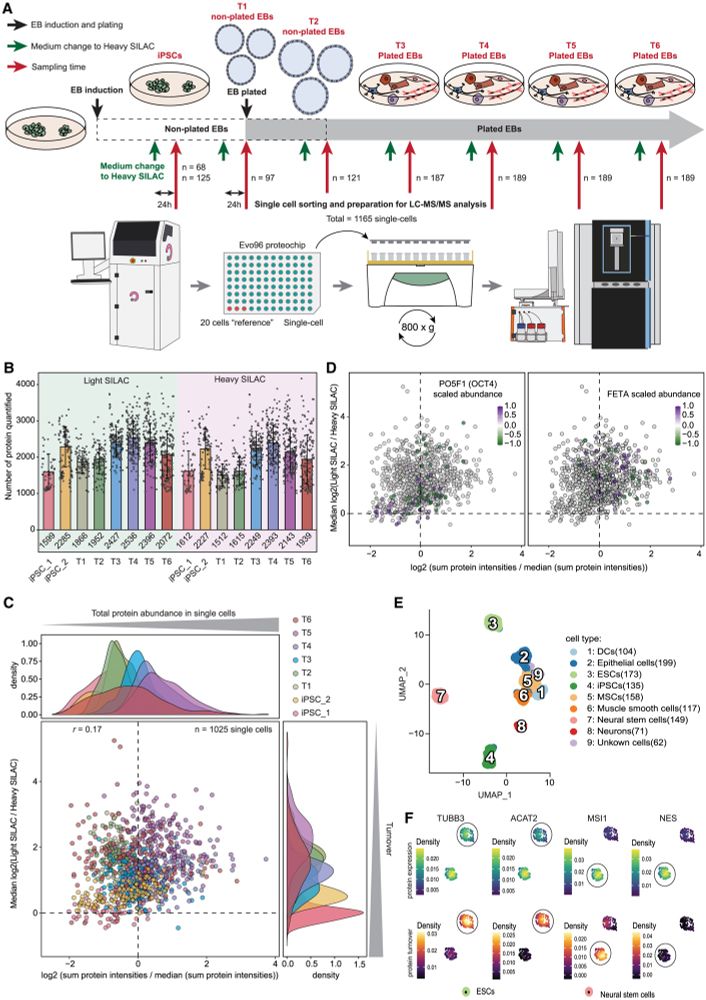
We then performed a large-scale SC-pSILAC experiment to study the undirected differentiation of human induced pluripotent stem cells through embryoid body induction in >1000 individual single cells.
Our comprehensive dataset shows protein turnover dynamics during cell differentiation over 2 months.
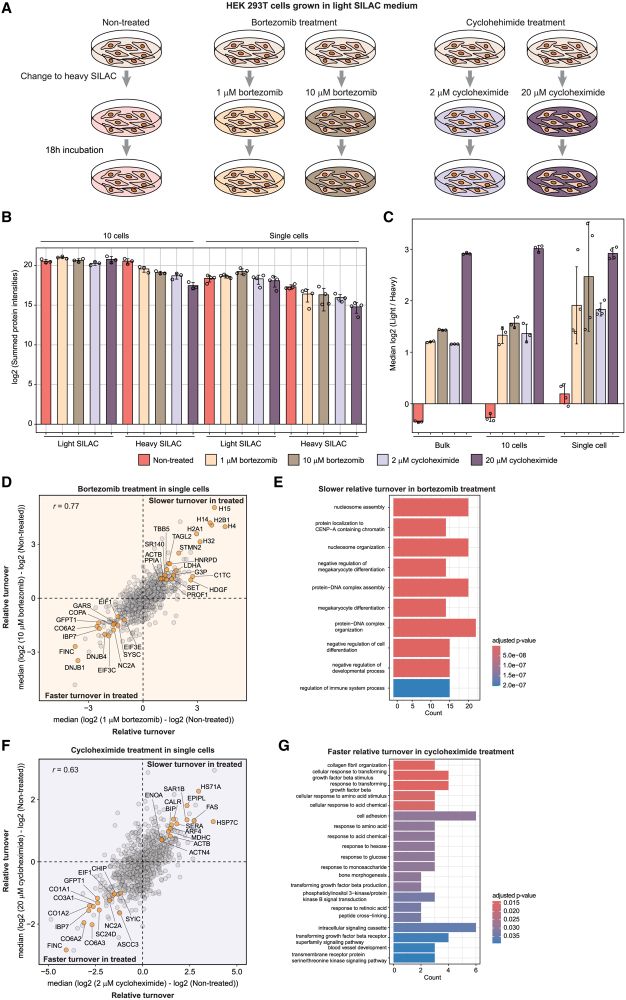
We first studied treatments altering global protein turnover including proteasome(bortezomib), protein translation (cycloheximide), and beta-cathenin inhibitions for altered turnover of a specific protein(GSK3 inhibitor).
SC-pSILAC revealed treatment-related changes in single cells.
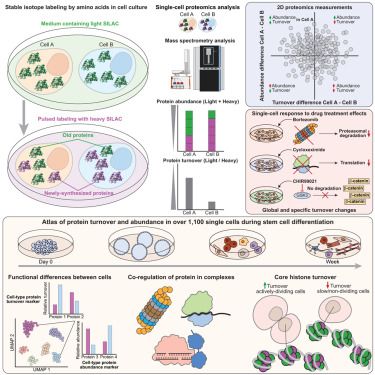
🚀 Excited to share our latest article in #singlecell proteomics published in Cell!
We've developed SC-pSILAC to simultaneously measure protein turnover and abundance in single cells, unlocking the first large-scale, 2D proteomic insights at single-cell resolution!
www.cell.com/cell/fulltex...
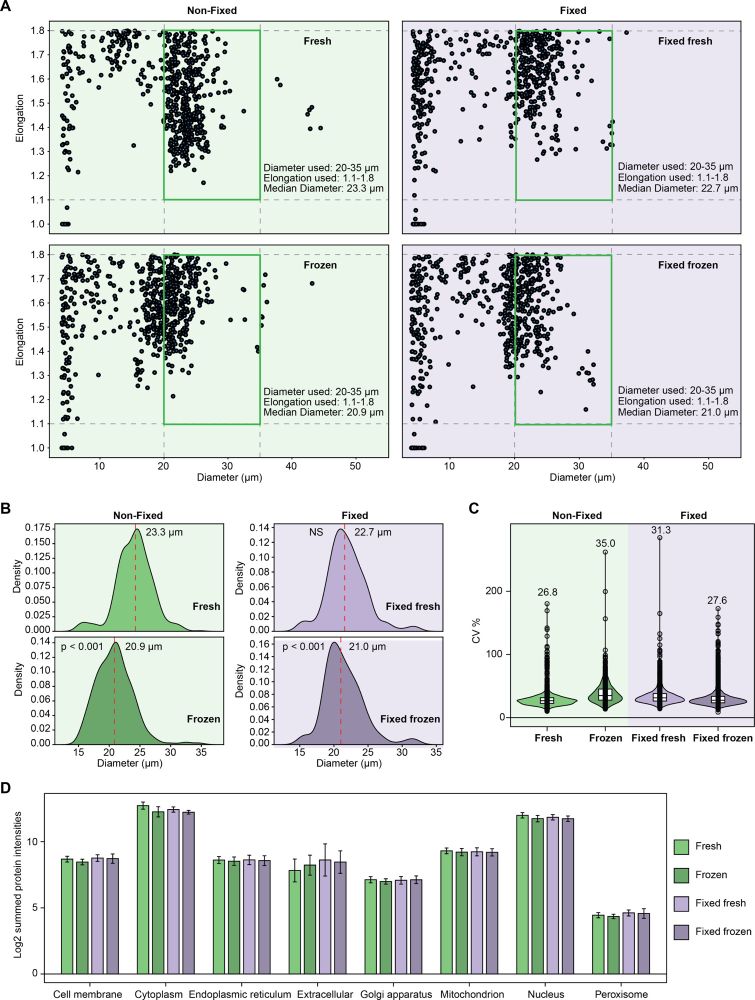
Finally fixed cells showed altered morphology vs. non fixed cells. Yet, after freezing the morphological changes in fixed cells are reduced compared to non fixed cells. Fixed frozen single cells have significantly lower CVs than non fixed frozen cells, enhancing data quality and reproducibility.
04.02.2025 08:15 — 👍 0 🔁 0 💬 0 📌 0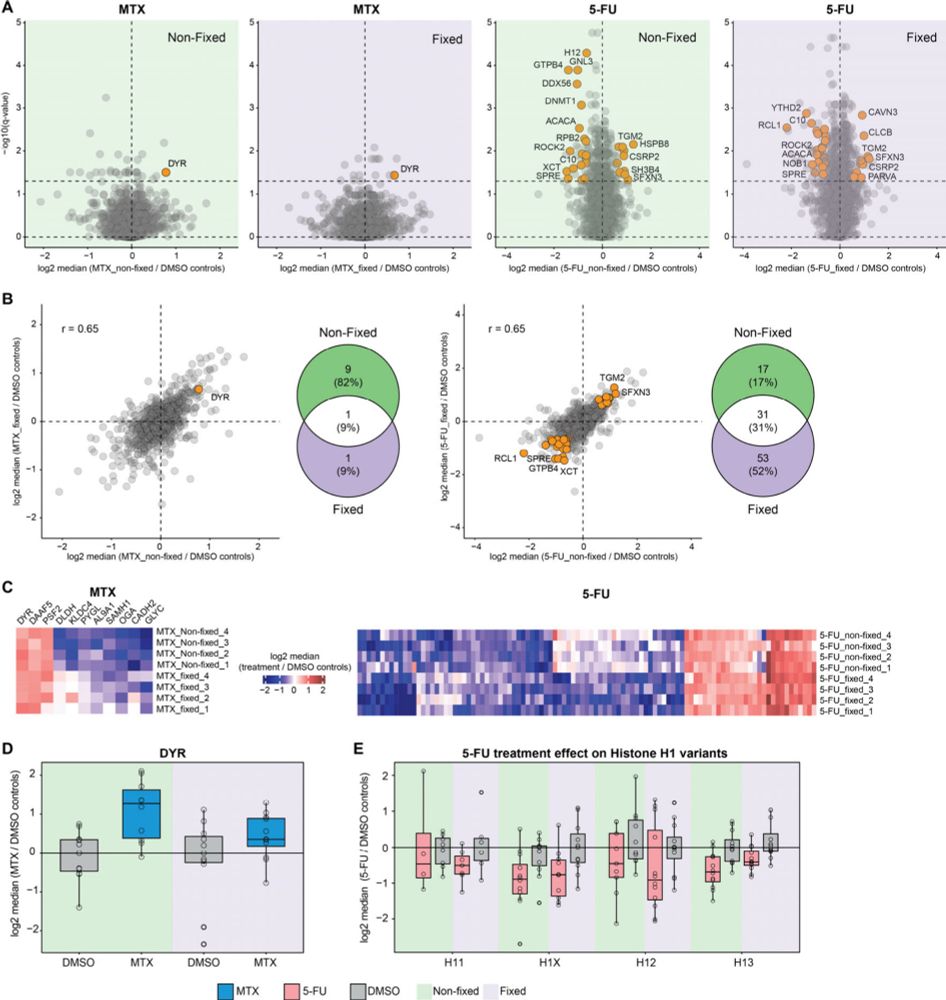
We treated 20 cells and single cells with 5-FU and MTX and demonstrated that fixation doesnt affect drug induced changes. While 20 cells provide deeper coverage single cell analysis still identifies key protein targets proving fixation is viable for SCP.
04.02.2025 08:15 — 👍 1 🔁 0 💬 1 📌 0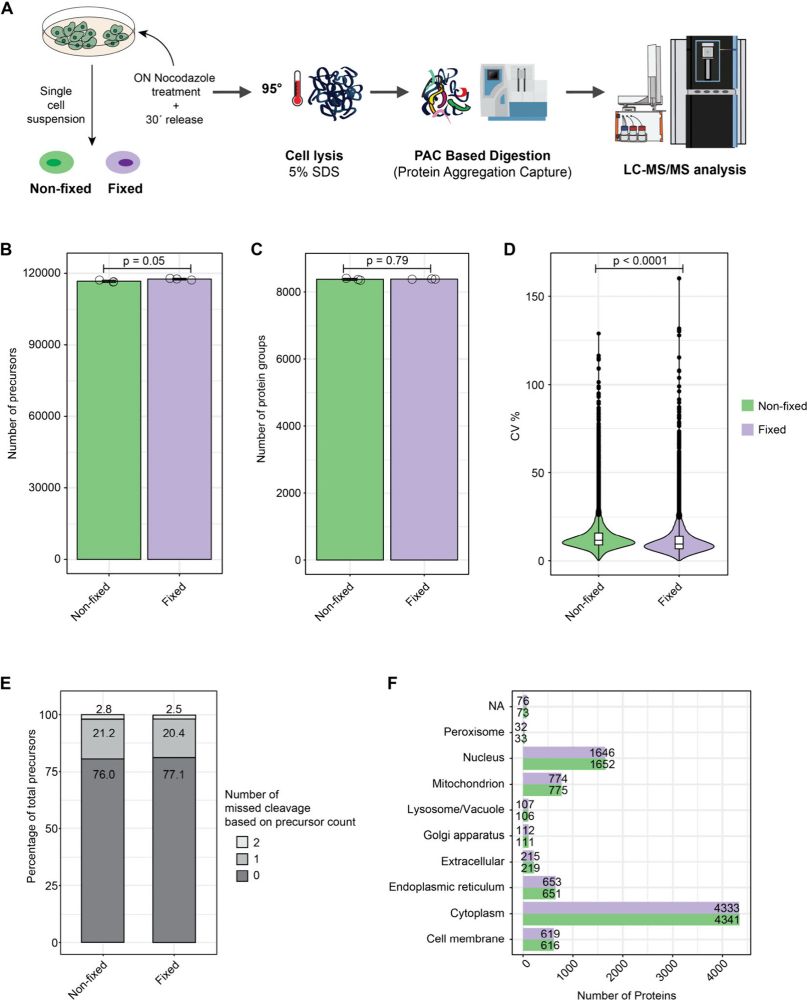
We tested bulk, OneTip and single cell proteomics with 2% formaldehyde and we found: no major effect on protein recovery, limited effect on digestion efficiency, lower CVs, and unaffected multiplexing potential.
04.02.2025 08:15 — 👍 0 🔁 0 💬 1 📌 0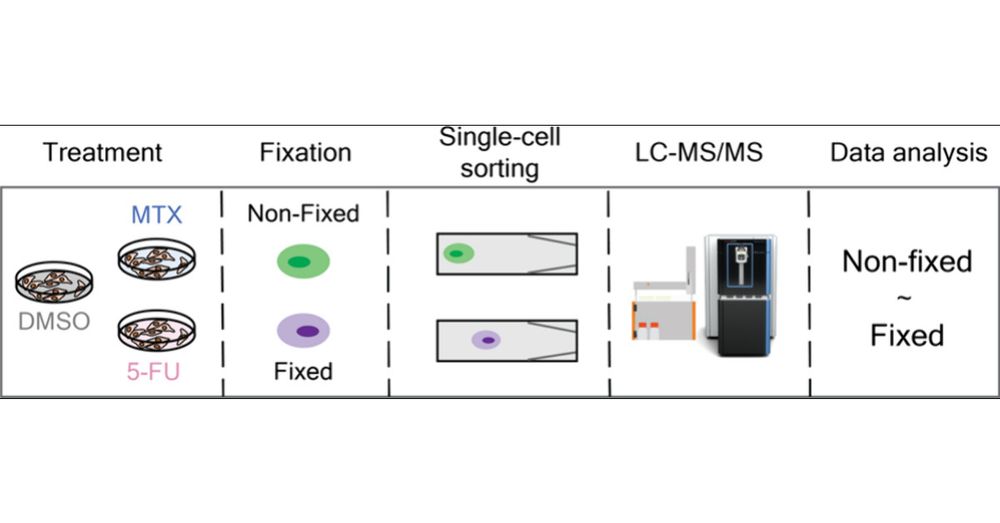
🚨 More research on single-cell proteomics!
We evaluated how formaldehyde-based fixation preserve proteome state, drug response and cell integrity. Ultimately, cell fixation can facilitate access to #singlecell by enabeling sample shiping and prolonged sorting 📦
pubs.acs.org/doi/10.1021/...
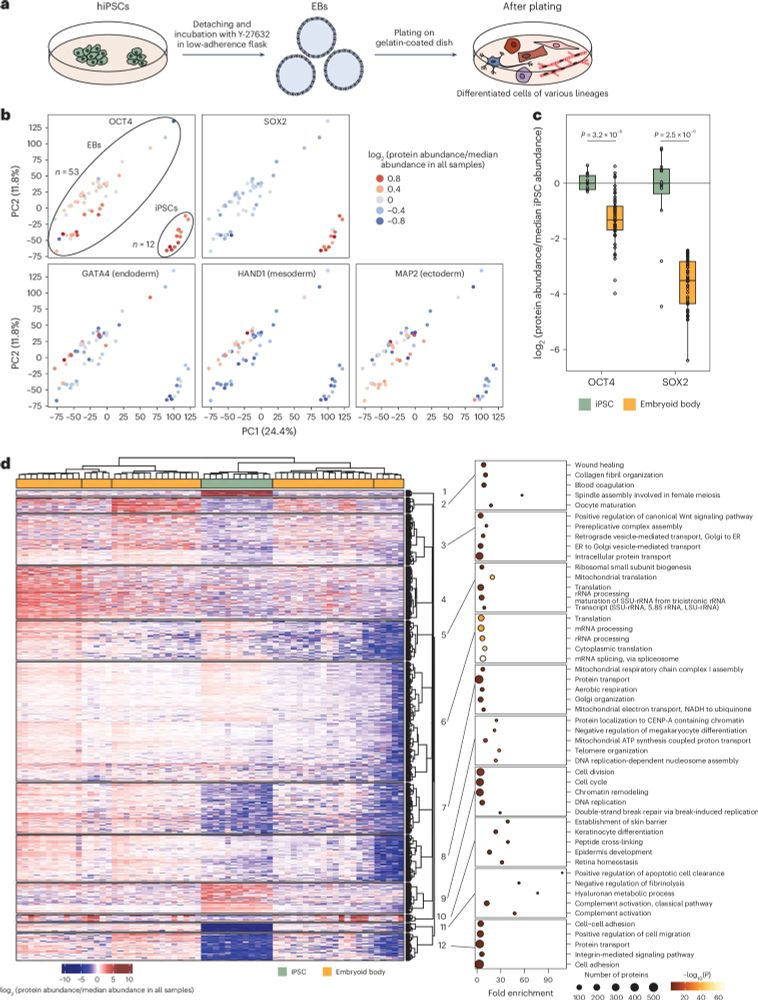
Lastly we applied the workflow to analyse undirected stem cell differentiation enabling the accurate quantification of stem cells and lineage markers.
17.01.2025 07:54 — 👍 1 🔁 0 💬 0 📌 0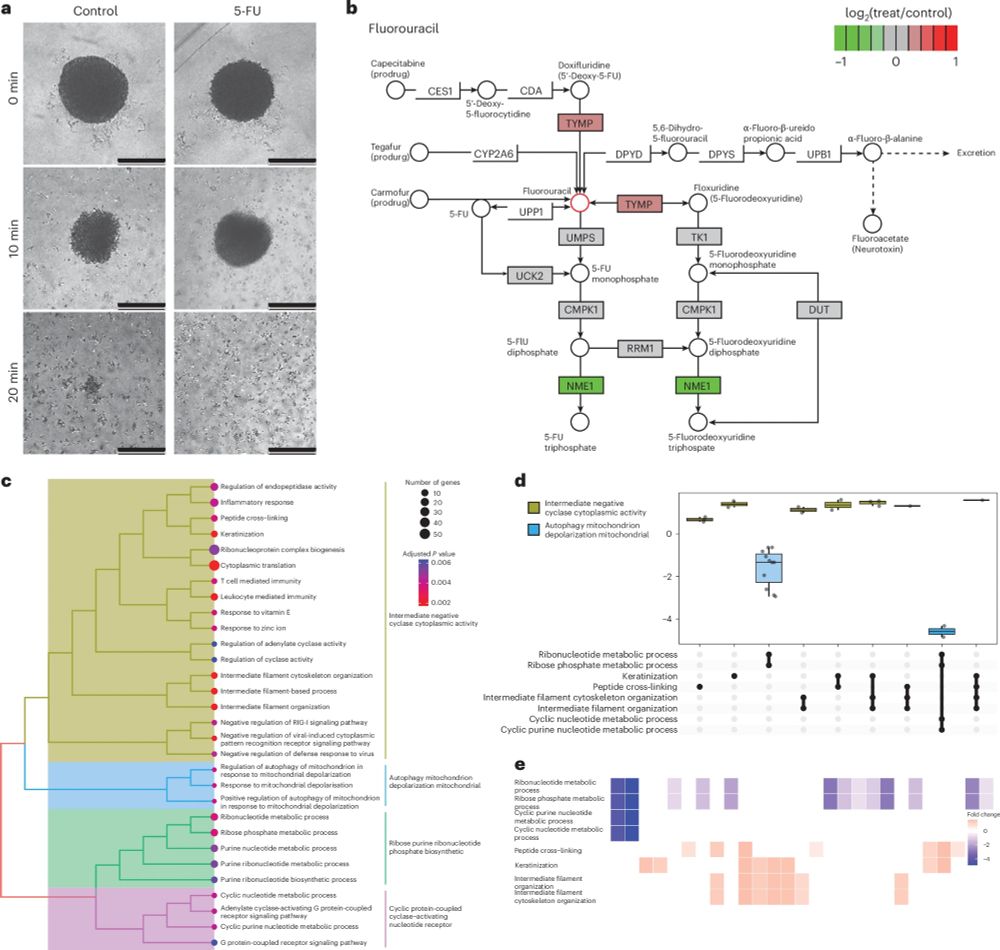
We further investigated the effect of 5-FU treatments on spheroids with the whisper 80SPD gradient, highlighting differential regulation of some 5-FU targets.
17.01.2025 07:54 — 👍 0 🔁 0 💬 1 📌 0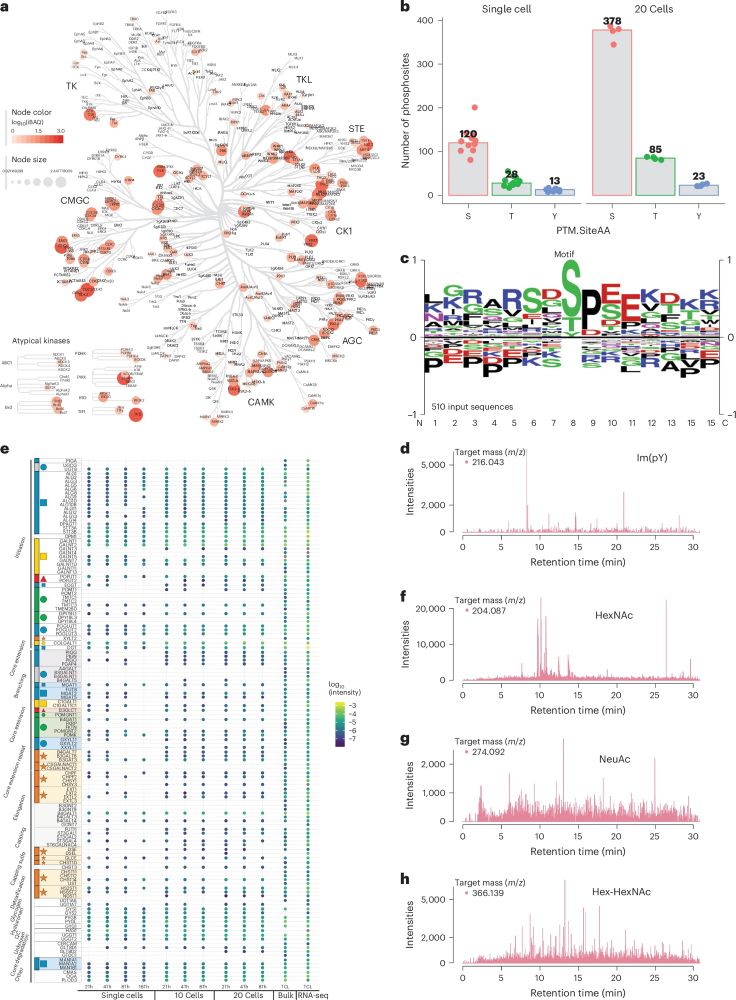
The improved sensitivity and peptide coverage allows for the detection of abundance PTM sites harboring phosphorylations and glycosylation without prior enrichment, opening new opportunities for SCP analysis.
17.01.2025 07:54 — 👍 1 🔁 0 💬 1 📌 0