May Institute on Computation & Statistics for MS and Proteomics is taking great shape. Fantastic new and repeat instructors and lots of new content. We are grateful to our sponsors Fragmatics and Biognosys. Keep an eye on the website computationalproteomics.khoury.northeastern.edu
10.11.2025 16:39 — 👍 22 🔁 9 💬 0 📌 0
Grateful
29.10.2025 14:06 — 👍 9 🔁 0 💬 1 📌 0
Very much looking forward! Thank you for the invitation 🙂
21.09.2025 23:20 — 👍 3 🔁 0 💬 0 📌 0
Yes!
07.07.2025 17:55 — 👍 2 🔁 0 💬 0 📌 0

This selfie was inspired by Livia Schiavinato Eberlin’s Biemann Award Lecture. #ASMS2025 @olgavitek.bsky.social
03.06.2025 21:49 — 👍 18 🔁 1 💬 0 📌 0

The Vitek lab is coming to #ASMS2025 in full force! Find us in the short courses, oral and poster sessions to talk about open-source statistical methods and software for MS proteomics and imaging!
30.05.2025 19:14 — 👍 20 🔁 2 💬 0 📌 0
May Institute – Computation and statistics for mass spectrometry and proteomics
A few more spots are still open for some of the in-person programs at May Institute on Computation and Statistics for Mass Spectrometry and Proteomics on April 28 – May 11, 2025 on the campus of Northeastern University in Boston MA computationalproteomics.khoury.northeastern.edu
15.04.2025 18:10 — 👍 16 🔁 9 💬 0 📌 1
What a fantastic recognition of a fantastic scientist. Ileana is an inspiration and a role model. Many, many congratulations
26.02.2025 17:06 — 👍 15 🔁 0 💬 1 📌 0
Please join us! There is still time to apply
25.02.2025 19:59 — 👍 7 🔁 2 💬 0 📌 0


USHUPO lightning talks session lightens the mood with @proteomicsnews.bsky.social @jyates.bsky.social Parag Mallick and many other incredible talents
24.02.2025 23:13 — 👍 21 🔁 3 💬 1 📌 0

My Book Is Out! Why I Wrote It and How You Can Help
Bridging the Gap Between Deep Learning and Causal Inference—A Code-First Approach
newsletter.altdeep.ai/p/my-book-is... The connection between genAI and causality is obvious but could nevery find any good learning material that made the connection.
So I wrote a book
24.02.2025 22:57 — 👍 9 🔁 2 💬 0 📌 0

Join us for the 2025 Barry L. Karger Medal Celebration! 🎉 March 10, 12–6 PM at Northeastern University, Boston. Hear from honoree Dr. Bernhard Küster & experts on biopharma, proteomics & systems biology. Don’t miss it! #Proteomics
Moderated by @olgavitek.bsky.social
19.02.2025 20:15 — 👍 5 🔁 2 💬 0 📌 0
💜
19.02.2025 12:03 — 👍 1 🔁 0 💬 0 📌 0
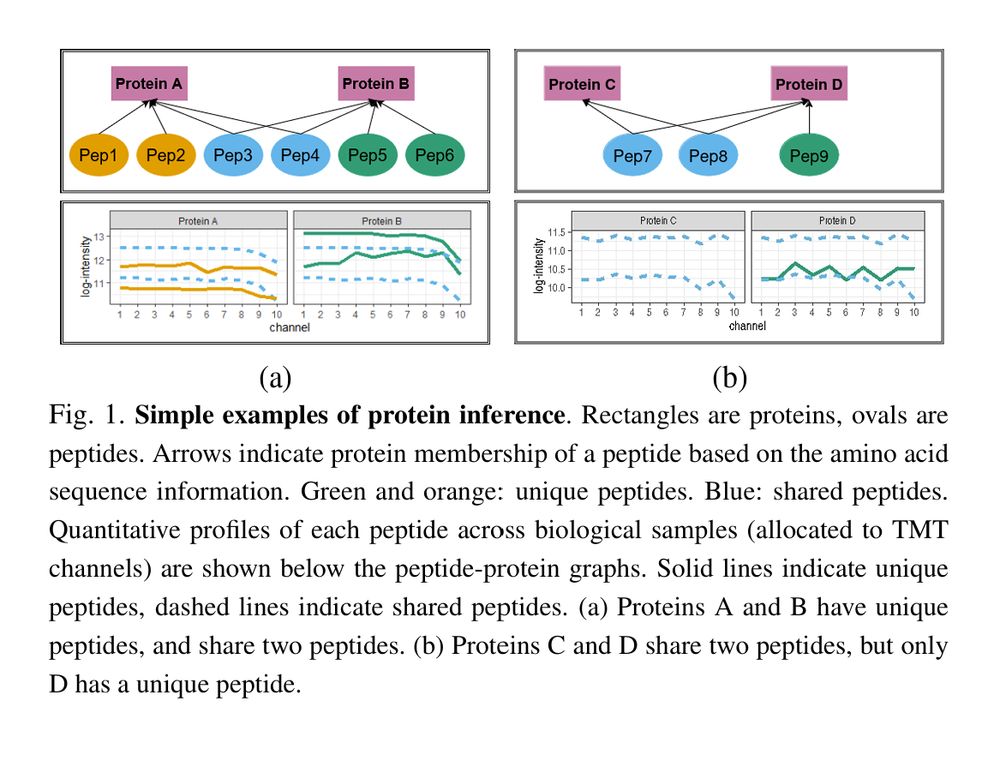
Relative quantification of proteins and post-translational modifications in proteomic experiments with shared peptides: a weight-based approach academic.oup.com/bio...
---
#proteomics #prot-paper
31.01.2025 21:20 — 👍 14 🔁 3 💬 0 📌 1
HUPO-PSI Spring Meeting 2025 – HUPO Proteomics Standards Initiative
🚨 Big news, #TeamMassSpec! 🚨
How do we unlock proteomics data reuse, tackle metadata challenges, and harness public (clinical) data for AI?
Find out at HUPO-PSI Spring Meeting 2025!
📅 March 31 – April 3, 2025
📍 Tübingen, Germany
(1/4)
07.01.2025 15:50 — 👍 15 🔁 7 💬 1 📌 0
May Institute – Computation and statistics for mass spectrometry and proteomics
May Institute on Computation and Statistics for Mass Spectrometry and Proteomics @KhouryCollege @Northeastern in Boston MA is happening in person on April 28-May 11, 2025 and is accepting applications! You will not regret attending computationalproteomics.khoury.northeastern.edu
12.01.2025 16:50 — 👍 50 🔁 28 💬 1 📌 1
@ypriverol.bsky.social Thank you for the shoutout to MSstats. Really wish it’d be mentioned in the manuscript too…
09.12.2024 23:10 — 👍 2 🔁 0 💬 0 📌 0
This is different from Covid testing because each protein has its own context-specific variation that needs to be characterized. In Covid you are only looking for one thing, and that variation can be characterized in advance.
06.12.2024 12:23 — 👍 1 🔁 0 💬 1 📌 0
Not a good idea because you lose all sense of biological variation. But you could create several pools of distinct samples from each condition and proceed with a comparison of the pools.
06.12.2024 12:21 — 👍 3 🔁 1 💬 3 📌 0
Somalogic, Olink, and Alamar are casting doubt on each other’s data, proudly calling out lack of correlation.
I know why they do this but in the long run as it casts doubt on affinity approaches as a whole.
We will think lots of data is generated that is trash and won’t know which
20.11.2024 22:42 — 👍 26 🔁 5 💬 4 📌 4
6th ESCP 2025 – APMA
It gives me great pleasure to announce our next European Single Cell Proteomics Conference www.apma.at/6thescp/
We would be delighted to receive many abstracts from students so that they have the opportunity to give their first presentation.
On behalf of: Erwin, Fabian, Manuel, Fabian and Karl
21.11.2024 07:14 — 👍 28 🔁 14 💬 0 📌 3
Hello world 😀
16.11.2024 22:01 — 👍 2 🔁 0 💬 1 📌 0
https://metabolomics.ucdavis.edu/
#metabolomics #bioinformatics #massspec #biochemistry #lipidomics
Warren Buffet Parody Page for Entertainment Purposes ONLY
Not a financial advisor 🚀 WSB 💴
First AI Bioinformatician built for Proteomics and beyond. Fast, intuitive analysis for scientists. Built by top-tier former Google/Verily engineers. Check us out at www.tesorai.com.
Postdoc Fellow at Saez-Rodriguez and Savitski labs (EMBL). Original from Córdoba, Spain.
Dietrich Volmer · Research in analytical chemistry, mass spec and metabolomics · Editor-in-Chief of Anal. Sci. Adv. + Editor of Rapid Commun. Mass Spectrom · Views my own
Humboldt University · Berlin · 🇨🇦🇩🇪 · volmerlab.de
Oficial Bluesky account of the Eastern North American Region of the International Biometric Society.
Subscribe to www.lawdork.com for SCOTUS, Trump, LGBTQ, criminal justice, and other legal news. / Email: lawdorknews@gmail.com / Signal: crg.32 / About me: Sober. Queer. Bipolar. Buckeye. / He/him.
Design engineer playing with AI and hacky prototypes @githubnext.com
Adores digital gardening, end-user development, and embodied cognition. Makes visual essays about design, programming, and anthropology.
📍 London
🌱 maggieappleton.com
Prof (CS @Stanford), Co-Director @StanfordHAI, Cofounder/CEO @theworldlabs, CoFounder @ai4allorg #AI #computervision #robotics #AI-healthcare
Assistant Professor in CS + AI at USC. Previously at Stanford, CMU. Machine Learning, Decision Making, AI-for-Science, Generative AI, ML Systems, LLMs.
https://willieneis.github.io
Developing data intensive computational methods • PI @ Seoul National University 🇰🇷 • #FirstGen • he/him • Hauptschüler
Scientist, Assistant Professor at MIT biology, #FirstGen
Assistant Professor, University of Toronto.
Junior Research Fellow, Trinity College, Cambridge.
AI Fellow, Georgetown University.
Probabilistic Machine Learning, AI Safety & AI Governance.
Prev: Oxford, Yale, UC Berkeley, NYU.
https://timrudner.com
Professor of Computational & Systems Biology at University of Manchester. Applies machine learning and modelling to help understand the complexity of biology. idsai.manchester.ac.uk, https://www.ellismcr.org, @official-uom.bsky.social
Professor, Santa Fe Institute. Research on AI, cognitive science, and complex systems.
Website: https://melaniemitchell.me
Substack: https://aiguide.substack.com/
Parent, spouse, Australian, Professor of Machine Learning in Oxford. Long Covid, trans rights, music, reggae on Fridays, AI must be good for humans, https://www.robots.ox.ac.uk/~mosb
Safe and robust AI/ML, computational sustainability. Former President AAAI and IMLS. Distinguished Professor Emeritus, Oregon State University. https://web.engr.oregonstate.edu/~tgd/