
🎖️Toutes nos félicitations à Sylvie Rétaux, cheffe de l'équipe DECA à NeuroPSI, qui a été nommée au grade de Chevalier de la Légion d’Honneur !
18.01.2026 20:18 — 👍 12 🔁 2 💬 0 📌 1@zenlabpasteur.bsky.social

🎖️Toutes nos félicitations à Sylvie Rétaux, cheffe de l'équipe DECA à NeuroPSI, qui a été nommée au grade de Chevalier de la Légion d’Honneur !
18.01.2026 20:18 — 👍 12 🔁 2 💬 0 📌 1
The call for abstracts is open to participate in the Ascona conference on "Neurogenesis from development to adulthood in health and disease" !
04.12.2025 19:53 — 👍 0 🔁 0 💬 0 📌 0
📣 Register to the 13th Advances in Stem Cell Biology Course #ASCBcourse at the @pasteur.fr @pasteuredu.bsky.social. Hands-on training in different models, embryonic stem cells, iPS cells and much more on #StemCells, #organoids. 🕘June22-July03, 2026
👉 shorturl.at/PyUHi




Great science & fruiful discussions last Friday the 14th Nov. around "the physics of Stem cells" , the 2025 Stem Cell Initiative & QBio joint symposium.
An honour to have Meryem Baghdadi & David Brückner as keynote speakers, who gave inspiring talks.
🙏🙏 to all our speakers & poster presenters.




⌛1 month left to apply – 2025 PPU call for candidates
We invite you to discover 4 Develpmental & Stem Cell Biology projects and 3 Immunology projects👇
🕰️Application deadline: October 20, 2025
📍Program starts: October 2026
Discover all the projects (50+) & apply: www.pasteur.fr/en/education...
We (with @nicolasdray.bsky.social @zenlabpasteur.bsky.social) are happy to release officially FishFeats, our #napari plugin to streamline 3D smFish/RNA quantification! 🎊
Available here: github.com/gletort/Fish... user documentation: gletort.github.io/FishFeats/

Pathway to Independence – an interview with Joaquín Navajas Acedo.
In this interview, @mads100tist.bsky.social talks about about his research vision, zebrafish and the importance of community:
journals.biologists.com/dev/article/...
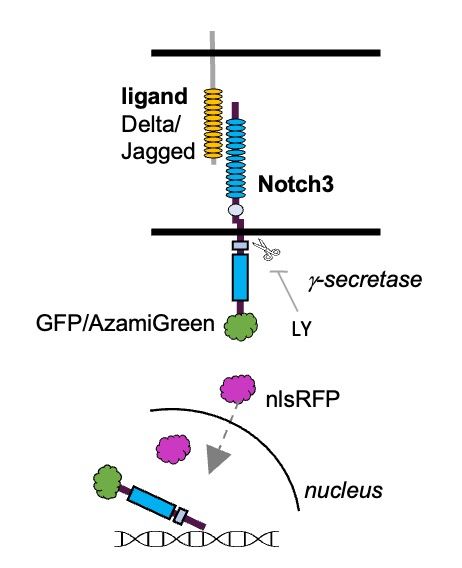
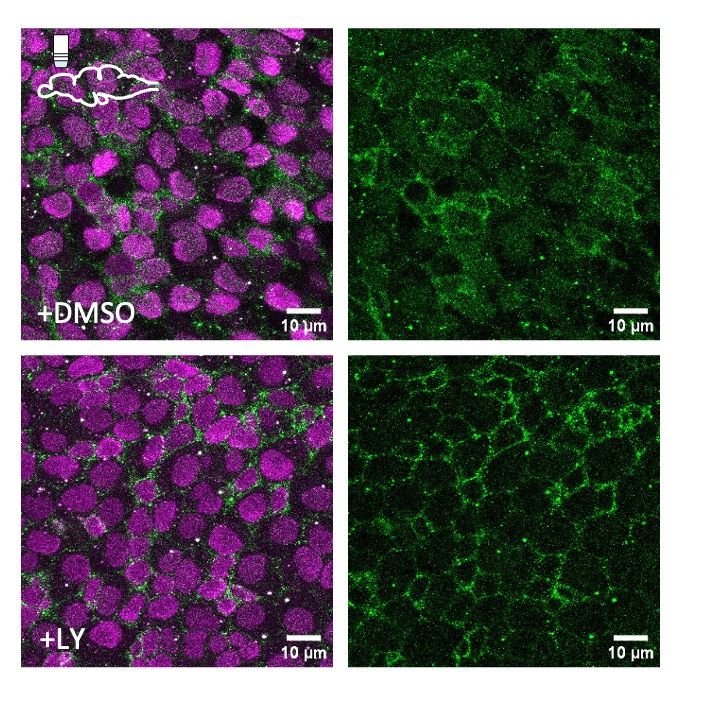
Translocation of the Notch3 Intracellular Domain (green) to the nucleus in adult neural stem cells (top), but retention at the membrane upon gamma-secretase inhibition (bottom).
New preprint out! Novel transgenic lines directly read out Notch3 signaling in situ, and reveal how adult neural stem cells coordinate quiescence and stemness by integrating spatiotemporal information from Delta and Jagged ligands. Read more: doi.org/10.1101/2025...
05.08.2025 13:57 — 👍 5 🔁 2 💬 0 📌 0

Engineer position, 12 months starting Oct. 1st, 2025, to study the role of NOTCH3 in human iPSC-derived neurogenesis models. Solid previous expertise required in cell culture, notably iPSCs, and organoid generation. Send CV, motivation letter and reference letter(s) to laure.bally-cuif@pasteur.fr
11.07.2025 20:21 — 👍 7 🔁 4 💬 1 📌 0
New call for research Group Leaders at Institut Pasteur Paris !
08.12.2024 19:36 — 👍 1 🔁 0 💬 0 📌 1
Just published in Development (@biologists.bsky.social):
doi.org/10.1242/dev....

On Mon 18/11, we held a mini symposium on the novel perspectives in human embryo research @institutpasteur. Lively discussions thanks to our prominent speakers/chair
Thanks to Nicolas Rivron; Alain Chédotal; Cécile Martinat; Ken McElreavey; Bernadette De Bakker and the chair Alfonso Martinez Arias

This also draws yet another parallel between physiological regulation stem cell pools and oncogenesis, which has been referred to as a process through which cells become “asocial” and cease to coordinate with their neighbors (18/18)
17.11.2024 20:09 — 👍 3 🔁 0 💬 0 📌 0
Although we were not able to identify what advantage this confers for NSC homeostasis, this non-canonical maintenance of Notch effectors illustrates different scales of quiescence regulation with some cells seemingly being able to listen less to their neighbors (17/18)
17.11.2024 20:08 — 👍 2 🔁 0 💬 0 📌 0
In vivo experiments confirmed that expression of nr2f1b in zebrafish NSCs is associated with a weaker response to Notch inhibition and functional experiments with morpholinos or electroporations of nr2f1b confirmed its ability to decrease sensitivity to Notch inhibition. (16/18)
17.11.2024 20:08 — 👍 1 🔁 0 💬 0 📌 0
Interestingly NR2F1 had previously been identified as a regulator of Notch signaling in the mouse inner ear (journals.biologists.com/dev/article/...), with downregulation of Notch effectors in NR2F1 knock-out mutants (15/18)
17.11.2024 20:08 — 👍 2 🔁 0 💬 0 📌 0
Using a gradient boosting method derived from SCENIC applied to both control and treated cells, we identified NR2F1/Coup-TF1 in sillico as a putative regulator enabling this resistance to Notch inhibition. (14/18)
17.11.2024 20:07 — 👍 1 🔁 0 💬 0 📌 0
Finally, we also identified that one cluster of cells barely responded to Notch inhibition, maintained expression of Notch effectors and did not upregulate genes associated with NSC activation. (13/18)
17.11.2024 20:07 — 👍 1 🔁 0 💬 0 📌 0
Although there are obvious differences between parenchymal astrocytes in mouse and astrocyte-like RGs in zebrafish, this suggests that comparative studies focusing on these cells might help better understand how to promote brain regeneration. (12/18)
17.11.2024 20:07 — 👍 1 🔁 0 💬 0 📌 0
The reported properties of the induced activation of these cells indeed shared similarities with that of astrocyte-like NSCs in the zebrafish pallium, such as participation in regeneration or reliance on Notch signaling (11/18)
17.11.2024 20:07 — 👍 1 🔁 0 💬 0 📌 0
Harmonizing the different reports of cell classification led us to consider that the cells that had been described as latent NSCs under different names in different publications were in fact likely the same population of parenchymal astrocytes close to the lateral ventricles. 10/18
17.11.2024 20:07 — 👍 2 🔁 0 💬 0 📌 0
Using the set of genes which we previously identified as allowing for distinction between murine RGL and astrocytes, we reclassified qNSC1 cells as astrocytes, in accordance with scRNAseq and in situ data from A Cebrian-Silla and methylome data from LPM Kremer. (9/18)
17.11.2024 20:06 — 👍 1 🔁 0 💬 0 📌 0
We found that the distinction between qNSC1 and qNSC2 had not been made in most cases even when the cells belonging to each group were present in the dataset. (8/18)
17.11.2024 20:06 — 👍 2 🔁 0 💬 0 📌 0
To get a clearer picture we re-analyzed scRNA-seq datasets encompassing the SVZ, generated by the labs of F Doetsch (@doetschlab.bsky.social), A Martin Villalba, J Frisen, S Linnarsson (@slinnarsson.bsky.social), J Biernaskie, N Rajewsky, F Miller, A Alvarez Buylla and O Raineteau. (7/18)
17.11.2024 20:05 — 👍 2 🔁 0 💬 0 📌 0
A similar phenomenon had been described in mouse astrocytes and putative subventricular zone (SVZ) radial glia-like (RGL) (called qNSC1) which respond to injury. (6/18)
17.11.2024 20:05 — 👍 1 🔁 0 💬 0 📌 0
In particular we found that one group of NSCs, recently described as being very similar to mouse parenchymal astrocytes, is efficiently recruited by Notch inhibition leading to the appearance of a cell state defined by co-expression of astrocytic and activating NSCs markers. (5/18)
17.11.2024 20:05 — 👍 1 🔁 0 💬 0 📌 0
With scRNAseq after 24hours of Notch inhibition we found that most cells had responded, but not in the same way. (4/18)
17.11.2024 20:04 — 👍 1 🔁 0 💬 0 📌 0
We attempted to push NSCs out of quiescence using Notch inhibition to see if previously identified clusters reacted differently to a pro-activation stimulus. (3/18)
17.11.2024 20:04 — 👍 1 🔁 0 💬 0 📌 0
Drawing from previous characterization of the heterogeneity of adult Neural Stem Cells (qNSCs) in the zebrafish pallium and the prediction that some of that heterogeneity comes from differences in quiescence depth (2/18)
17.11.2024 20:04 — 👍 1 🔁 0 💬 0 📌 0
New pre-print from the lab out on @biorxivpreprint (www.biorxiv.org/content/10.1...). 1/18
17.11.2024 19:51 — 👍 3 🔁 0 💬 0 📌 0