RE: https://fosstodon.org/@gicrisf/115515504598989643
that sounds useful! are there other #cheminformatics toolkits integrated into #Emacs ?
09.11.2025 09:43 — 👍 0 🔁 2 💬 0 📌 0

BChemXtract - a ChemDraw parser and structure extractor - is our first open source software project and is available on @github:
🔗 https://github.com/Beilstein-Institut/BChemXtract
#openscience #opensource #FAIRdata #FAIRchemistry #cheminformatics #GitHub #BeilsteinCheminfoLabs #BChemXtract
05.11.2025 14:00 — 👍 3 🔁 8 💬 0 📌 0
Original post on fosstodon.org
Euclid 2.13 is released: https://github.com/BlueObelisk/euclid/releases/tag/euclid-2.13
Minor update that is tested with Java 21 and 25, for which it removed the unused ThriftyList class. It also upgraded dependencies to Log4j 2.25.2 and Commons Lang3 3.19
Euclid is a library of numeric […]
02.11.2025 16:37 — 👍 1 🔁 0 💬 0 📌 0

What is Rogue Scholar? – Rogue Scholar Documentation
"Getting a DOI on personal blog posts is technically challenging, but possible using services such as The Rogue Scholar (https://docs.rogue-scholar.org/)." https://doi.org/10.1007/s44186-024-00287-w
12.10.2025 07:33 — 👍 8 🔁 11 💬 0 📌 0
#compchemsky
Now offers tools to save computed spectra as PNG, PDF, SVG .. or the underlying data.
Found a really nice #opensource #openscience plotting library
27.09.2025 17:04 — 👍 20 🔁 2 💬 1 📌 0
25 years of the Chemistry Development Kit
Twenty five years ago the Chemistry Development Kit (CDK) was founded. The Chemistry and Internet (ChemInt2000) had just ended (it ran from 23 to 26 September) and my friend and I had taken the Amtrak night train from Washington to South Bend. At that time there were two leading Java applets for chemistry, JChemPaint and Jmol. I had hacked Chemical Markup Language support into both of them, and Dan Gezelter (Jmol and openscience.org), Christoph Steinbeck (JChemPaint), and me took the opportunity of being in North America to discuss if we could use a common code base. Chris’ _compchem_ had done something similar. Peter Murray-Rust, who had also attended ChemInt2000 like me and Chris did not attend.
I do not remember exactly, but I guess we must have met on the 28th and 29th? Maybe already on Wednesday. During this meeting we discussed a common data model (yes, Jmol used the CDK data model at some point) and somewhere during the meeting we wrote down a name for the project. There was the Java Development Kit, so this could be the Chemistry Development Kit. The name stuck.
A quick post like this cannot do credit to the history of the CDK, nor of everyone involved in the past or still is. You can browse some of the history of the CDK in my blog and in Chris’ blog. It has been an amazing journey and with a small grant just behind us (with Alyanne de Haan, René van der Ploeg, and Marc Teunis from Hogeschool Utrecht), and all the awesome things ongoing (new JChemPaint, various extensions, upgraded downstream tools), the CDK is alive and kicking.
A huge congrats and thanks to everyone (and every company and organization) who contributed code to the CDK with this huge milestone. There are a few people that I want to particularly thank (see the AUTHORS file for all names): Chris, who in the late nineties made a difference with open source in chemistry, Dan, for Jmol and hosting this memorable meeting at Notre Dame University, Rajarshi Guha, who operated _CDK Nightly_ for many years, well before Travis and Google Actions, Stefan, Miguel, Gilleain, and Christian, for many years of contributions to the CDK, and John Mayfield, the current CDK release manager.
25 years Chemistry Development Kit (CDK) and going strong! :) https://doi.org/10.59350/4ce2c-fxh02 https://chem-bla-ics.linkedchemistry.info/2025/09/28/25-years-of-the-chemistry-development-kit.html
Replying to this post will make it show up in my blog.
#openscience #chemistry #opensource
28.09.2025 08:41 — 👍 3 🔁 8 💬 0 📌 0
this week, 25 years ago, @steinbeck, @egonw, and @gezelter came together in South Bend., USA and founded the Chemistry Development Kit
#chemistry #openscience
26.09.2025 10:30 — 👍 0 🔁 8 💬 1 📌 0

Screenshot of a frame of one of the videos in the linked pull request, showing a chemical structure with various R-groups on the left (code and R group values), and on the right a list of SMILES combinatorial structures generated from the input on the left.
nice new R-group SMILES enumeration feature in the upcoming JChemPaint release, see https://github.com/JChemPaint/jchempaint/pull/225
#openscience #chemistry
11.09.2025 11:40 — 👍 0 🔁 7 💬 0 📌 0
Original post on fosstodon.org
pybacting 0.2.15 is released with Bacting 1.0.7 (see also https://social.edu.nl/@egonw/115040533430981814): https://pypi.org/project/pybacting/0.2.15/
pybacting is a Python wrapper around Bacting which provides functionality from the Chemistry Development Kit, OPSIN, Oscar4, BridgeDb, InChI […]
23.08.2025 10:51 — 👍 0 🔁 2 💬 0 📌 0
Original post on fediscience.org
@mike
When #Elsevier sent takedown notices to a few #Harvard authors who posted copies of their articles to their personal web sites (2014), I did an interview with Harvard Library and replied much as you did […]
14.08.2025 15:37 — 👍 0 🔁 0 💬 0 📌 0
Original post on fosstodon.org
Oscar4 5.3.0 has been released: https://github.com/BlueObelisk/oscar4/releases/tag/5.3.0
"OSCAR (Open Source Chemistry Analysis Routines) is an open source extensible system for the automated annotation of chemistry in scientific articles. It can be used to identify chemical names, reaction […]
05.08.2025 10:32 — 👍 0 🔁 1 💬 0 📌 0

We have recently delivered around 40,000 chemical names to the One Million IUPAC Names project of Egon Willighagen @egonw. A CC0 license provides the community with a free and reusable set of chemical names.
We encourage the community to join us in supporting […]
[Original post on hessen.social]
17.07.2025 13:00 — 👍 1 🔁 5 💬 1 📌 0

2D chemical depiction of a chemical structure with nine sugar-like rings from the longest IUPAC name found so far, with 626 characters in length.
new: 'One Million IUPAC names #3: the 200 thousand milestone and 1 million IUPAC names" https://doi.org/10.59350/6f7he-kxt56 https://chem-bla-ics.linkedchemistry.info/2025/06/09/one-million-iupac-names.html
"I could not find the time earlier to report (reason) […]
[Original post on social.edu.nl]
09.06.2025 09:59 — 👍 0 🔁 6 💬 0 📌 0
Original post on fosstodon.org
new preprint with #opensource #cheminformatics by @Kohulan et al.: "Cheminformatics Microservice V-3: A Web Portal for Chemical Structure Manipulation and Analysis" https://doi.org/10.26434/chemrxiv-2025-xjkxl
"Here, we present Cheminformatics Microservice V3, a significant update to the […]
06.06.2025 07:51 — 👍 1 🔁 6 💬 0 📌 0
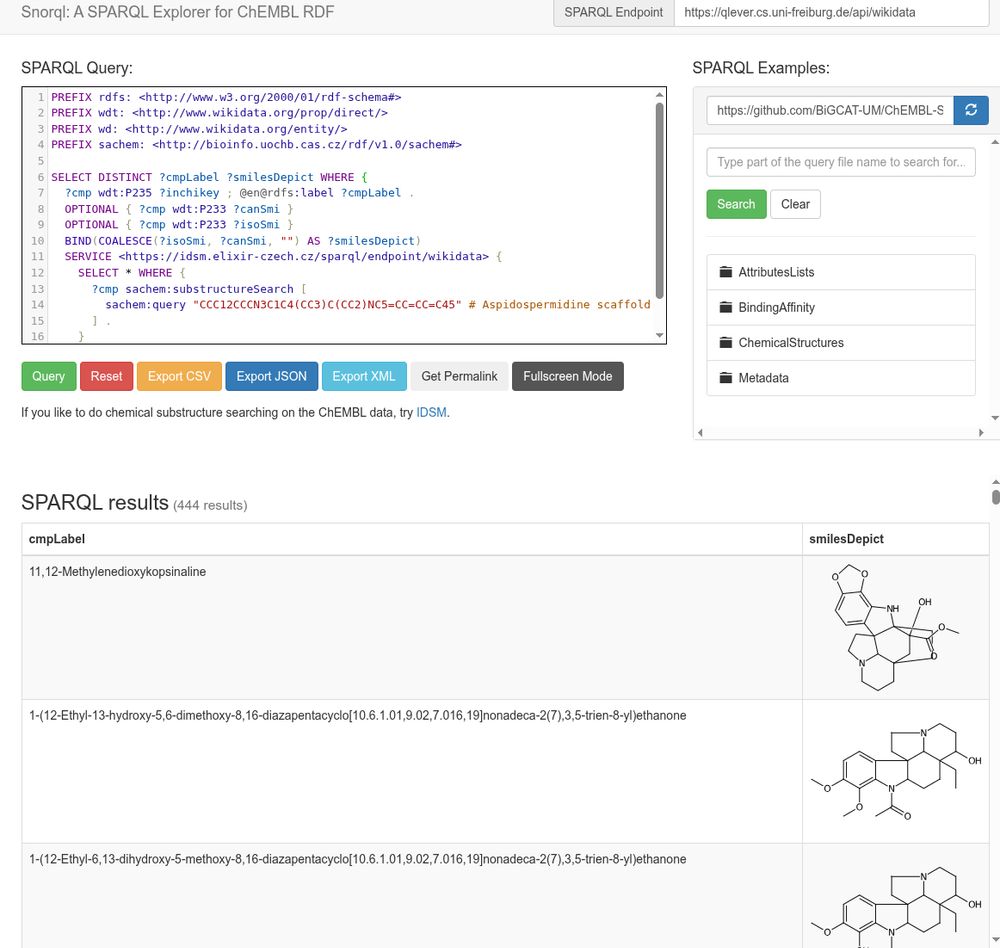
Screenshot of Ammar's Snorql UI, here wrapped around the Wikidata SPARQL instance by the QLever team, federating with the IDSM SPARQL endpoint for substructure searching, and using CDK Depict to convert the SMILES into 2D depictions in the results tables
oolonek reminded me how cool IDSM is (see https://doi.org/10.1186/s13321-021-00515-1) and works wonderfully in the rich chemistry SPARQL world (see https://doi.org/10.1186/1758-2946-3-15).
Here I mashed up @wikidata, IDSM, QLever, @cdk Depict, and Ammar […]
[Original post on social.edu.nl]
29.05.2025 10:16 — 👍 0 🔁 6 💬 0 📌 0
Original post on fosstodon.org
Fwd: "OPSIN's new home is at EMBL-EBI" https://chembl.blogspot.com/2025/05/opsins-new-home-is-at-embl-ebi.html
"We are delighted to announce that the OPSIN web app has become a member of the Chemical Biology Services here at EMBL-EBI and can be accessed at:
https://www.ebi.ac.uk/opsin " […]
16.05.2025 11:25 — 👍 0 🔁 6 💬 0 📌 0
the @intconfchemstr is held soon. At least two Blue Obelisk members are going to meet up. Who else likes to join?
#openscience #chemistry
11.05.2025 07:57 — 👍 0 🔁 6 💬 0 📌 0
Too bad you can't give a Nobel prize to the PDB. How much science and medicine has been enabled because of this open resource?
#OpenScience
08.05.2025 16:25 — 👍 35 🔁 12 💬 1 📌 0
Original post on fosstodon.org
The 2025_03_1 release of #RDKit release includes my contribution to speed up part of getting 2D fingerprints for a molecule by ~75x! So if you generate #chemical fingerprints, now is a good time to upgrade.
Reminder that I'm #OpenToWork so if you're hiring for #cheminformatics or […]
01.05.2025 15:11 — 👍 5 🔁 3 💬 1 📌 0
Original post on fosstodon.org
"Against common expectations, scientific projects have a longer lifetime than matched non-scientific open-source software projects. We expect our curated attribute-rich collection to support future research on scientific software and provide insights that may help extend longevity of both […]
29.04.2025 13:54 — 👍 1 🔁 2 💬 0 📌 0

Screenshot of the GitHub "commits over time" chart, showing the weekly number of commit, starting in February, and going down until mid March, when it stabilized at around 5 commits per day, each about ~1000 new IUPAC names. Until the week of April 21, then jumping to over 35 commits, reflecting a change in the workflow, as explained in the post.
new blog: "One Million IUPAC names #2: the 100 thousand milestone" https://doi.org/10.59350/dycsw-qeq51 https://chem-bla-ics.linkedchemistry.info/2025/04/27/one-million-iupac-names-2-the-100-thousand-milestone.html
"Two and a half month into the One Million […]
[Original post on social.edu.nl]
27.04.2025 13:56 — 👍 0 🔁 3 💬 0 📌 0
Original post on social.edu.nl
but then I also read news like this, which stems me optimistic: "The combined efforts of the recipients are anticipated to create almost 100,000 new quantitative reaction data points which will be published under a CC BY-SA license in the structured ORD format, ready for machine learning by […]
24.04.2025 08:46 — 👍 0 🔁 2 💬 0 📌 0
Original post on fosstodon.org
#openscience #cheminformatics dates back to the late nineties with the emerging collaborative development of JChemPaint, Jmol, and the Chemical Markup Language. Sketch of the history by Chris Steinbeck: "The evolution of open science in cheminformatics: a journey from closed systems to […]
05.04.2025 04:30 — 👍 0 🔁 4 💬 1 📌 0
Original post on fosstodon.org
CMLXOM 4.11 has been released: https://doi.org/10.5281/zenodo.15108779
"Minor release, reverting to (the newer) xml-apis 1.4.01, updating to Joda time 2.14, and removing unused imports, updating deprecated code, and minimal added JavaDoc."
CMLXOM is a Java library for reading and writing […]
30.03.2025 13:50 — 👍 0 🔁 1 💬 1 📌 0











