



🎄✨ Last Friday at #IBMP: a cozy Christmas celebration with homemade vin chaud and delicious mannele 🥨🍷 A perfect moment to share, laugh, and wrap up the year together. Science also means community!
10.12.2025 07:42 — 👍 1 🔁 0 💬 0 📌 0@ibmp-cnrs.bsky.social
IBMP is the largest @cnrs.fr center devoted to #plantscience in France | www.ibmp.cnrs.fr




🎄✨ Last Friday at #IBMP: a cozy Christmas celebration with homemade vin chaud and delicious mannele 🥨🍷 A perfect moment to share, laugh, and wrap up the year together. Science also means community!
10.12.2025 07:42 — 👍 1 🔁 0 💬 0 📌 0
🌱Great momentum at the TYPEX Days 2025 in Strasbourg: new Prime Editing tools, progress across crop species, and a growing shared cloning platform. More here 👉 www.pepr-selection-vegetale.fr/actualites/j...
09.12.2025 20:16 — 👍 3 🔁 2 💬 0 📌 0We are seeking a PhD student to investigate the deadenylation machinery in Arabidopsis seeds as part of the IMCBio program. @ibmp-cnrs.bsky.social @igbmc.bsky.social
If you are interested, please visit the following link (PhD-2026-30):
imcbio-phdprogram.unistra.fr

Reinforcing the 🇫🇷France–Japan🇯🇵 collaboration in plant biology🌱
Great meeting our French and Japanese colleagues in a cozy pre-Christmas atmosphere in Strasbourg, hosted by @ibmp-cnrs.bsky.social
So glad to have brilliant Japanese researchers at @rdplab.bsky.social
Let’s grow science together! 🌱✨

🎓 Spotlight on Judith Estefanía López Ponce, PhD student in Manfred Heinlein’s group (IBMP), within the ViroiDoc MSCA network!
🔗 linkedin.com/posts/viroid...
#ViroiDoc #MSCA #PlantScience

How plant mitochondria make their own proteins 🌱🔬
A new study published in @natcomms.nature.com reveals the structure of the cauliflower mitoribosome and a unique RsgA mechanism that blocks premature translation.
▶️https://nature.com/articles/s41467-025-65864-z
#PlantBiology #CryoEM #Ribosome
➡️ #PhDPosition #PhDOpportunity #PlantScience #PlantBiology #PlantMolecularBiology #Genetics #Epigenetics #CRISPR #Arabidopsis #ResearchJobs #WeAreHiring #JoinOurLab
02.12.2025 08:01 — 👍 2 🔁 1 💬 0 📌 0
🚀 IMCBio PhD Fellowships 2026 - 5 PhD in plant biology open at IBMP :
🌱 RNA biology
🧬 Translation
🔬 Chloroplast & mitochondria
🧫 Antiviral defence
🌾 Seed biology
📅 Apply before 25 Jan 2026
🔗 imcbio-phdprogram.unistra.fr
Join us and push the boundaries of plant science! 🌿



🦠PhD Defense🎓
👏Congrats to Lucas Schalck, who defended his PhD “Exploration fonctionnelle de la protéine P3a du Turnip yellows virus” supervised by Véronique Ziegler-Graff

Delphine Pott (left) and Marco Incarbone (right) standing in front of a yellow wall and next to a window facade in an office building. Both are smiling.
Great talk by @delphinem-p.bsky.social from @geminiteamlab.bsky.social yesterday: “Hijacking the Plant #Spliceosome” on how alternative splicing drives #viral infection and #tomato fruit development 🍅
Thanks to our host @incavirus.bsky.social for organizing this insightful seminar! 👏


Interreg KliwiResse project project completed :
➡️New resources generated, including metabolic stress markers identified at IBMP.
➡️Strong partner network built for future UpperRhine collaborations.
🔗 More info: kliwiresse.wine-science.eu/news_en.htm

Cool bonus from participating on the IMCBio event today @ibmp-cnrs.bsky.social is to attend to an amazing seminar by the one and only @pierremarcdelaux.bsky.social !, what an awesome coincidence!
17.11.2025 15:19 — 👍 10 🔁 1 💬 1 📌 0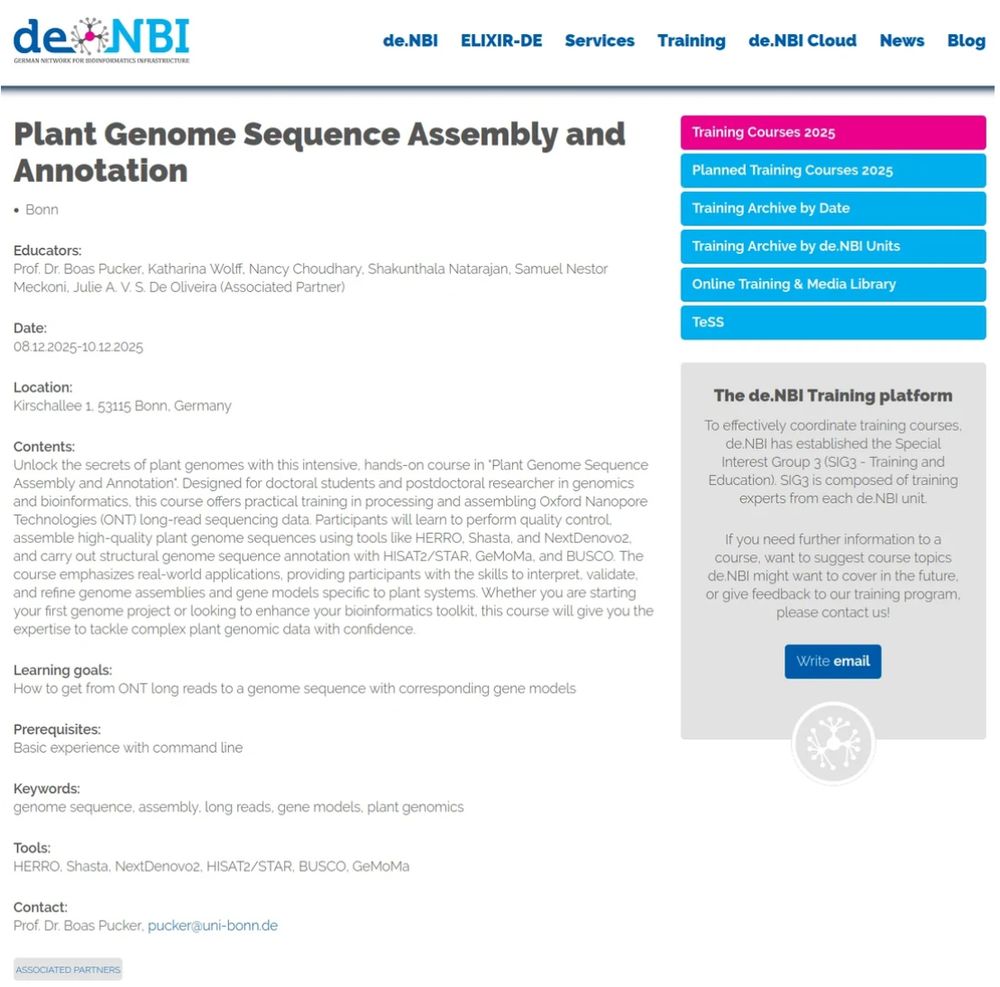
Genomics course announcement. Please find details here: https://www.izmb.uni-bonn.de/en/pbb/news#denbi2025
Curious about plant genomics? 🌿
Join our upcoming training courses to explore how plant genomes are assembled and annotated.
Details 👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #Bioinformatics #DeNBI #PlantScience
@denbi.bsky.social @puckerlab.bsky.social
Guillaume Hummel, David Pflieger, Alexandre Berr, Laurence Drouard: From the RNA world to land plants: Evolutionary insights from tRNA genes https://arxiv.org/abs/2511.01943 https://arxiv.org/pdf/2511.01943 https://arxiv.org/html/2511.01943
05.11.2025 06:50 — 👍 2 🔁 2 💬 0 📌 0
🦠PhD Defense🎓
Congrats to Amélie Janzam, who defended her PhD “Role of the non-coding RNA3 and the p14 protein in long-distance viral movement and RNA interference suppression of BNYVV.” Supervised by David Gilmer
#PhDDefense #PlantVirology #PlantBiology

🧬PhD Defense🎓
👏Congrats to Margaux Cheminat, who defended her PhD on “Deciphering the molecular interaction between Streptomyces and barley: influence of the brassinosteroid signaling pathway.”
Supervised by Florence Arsène-Ploetze & Hubert Schaller
#PlantBiology #Microbiology #PhDDefense

Thrilled to share our new preprint on the evolution of transfer RNA (tRNA) genes in photosynthetic organisms, together with David Pflieger, Alexandre Berr, and Laurence Drouard!
#PlantGenomics #Evolution #tRNA
@ibmp-cnrs.bsky.social
arxiv.org/abs/2511.01943

The 1st chapter of the PhD project of
@rachnabehl.bsky.social became online as pre-print. A meta-analysis of epigenomic changes in plants under environmental stresses…
#PlantScience #Epigenetics
www.biorxiv.org/content/10.1...

🌱 Light does more than fuel photosynthesis, it guides plant growth and survival. Discover how the TOR kinase connects light signals to metabolism and stress responses in a new
@jxbotany.bsky.social review featuring Mikhail Schepetilnikov
#TEsky #PlantScience 🌾🧪
16.10.2025 21:11 — 👍 4 🔁 3 💬 0 📌 0
I’m very pleased to share our latest work, led by the talented @cbuckley.bsky.social as part of his PhD. It’s our first attempt at studying #circadian rhythms in wheat with the help of some great collaborators @jesshyles.bsky.social @adaevo.bsky.social et al. #plantsci 🌾⏰ 1/7
doi.org/10.1111/nph....

Congratulations to Jean-Michel Davière and Stefan Grob, laureates of the 2025 IDEX call from the @unistra.fr 🎉
A great recognition of their innovative projects and the excellence of research at IBMP! 🌱🔬

On Oct 7, IBMP group leaders visited the Joseph Gottlieb Kolreuter Institute (JKIP) at @kit.edu. A great day of science, discussions, and new connections — from inspiring talks by Holger Puchta & Natalia Requena to a visit of the Karlsruhe Botanical Garden and greenhouses. 🌱
16.10.2025 10:26 — 👍 3 🔁 2 💬 0 📌 0



A great day at #Mifobio2025! 🌱🔬
From seed germination to Arabidopsis leaves and zebrafish hearts, our team explored the fascinating diversity of living systems with the EnderScope, pushing the limits of live imaging during this inspiring edition !
@cnrs-alsace.bsky.social @igbmc.bsky.social

🧪 Journée de cohésion des personnels des plateformes de l’IBMP à l'IS2M ! Une journée enrichissante autour des savoir-faire techniques et scientifiques de nos plateformes respectives.
Un grand merci à l’équipe de l’IS2M pour son accueil 👏
@cnrs-alsace.bsky.social #ScienceEnPartage

Avec le CNRS en Alsace 2023-2024
Le rapport d'activité 2023-2024 du CNRS Alsace est paru !
Partage des savoirs, innovation, sciences & recherche, témoignages, Europe & international... Un contenu à picorer ou à lire de bout en bout. Bonne lecture !
➡️ www.alsace.cnrs.fr/fr/rapport-d...
@cnrs.fr


🔎 “Tolerance: the plants that make peace with viruses” 🌿 🦠
An activity presented by @ibmp-cnrs.bsky.social, as part of the Science with and for Society program of the French National Research Agency (ANR). Visual support produced by CNRS Alsace.

#BDécouvrons SynPPR
Un "scratch" à ARN pour bloquer la production de protéines indésirables en #agriculture et #santé humaine ? 🌽🩺
C'est le défi que relèvent Kamel Hammani et son équipe de l'IBMP.
BD par Camille Van Belle 👉 bit.ly/synppr-infos
#SAPS_CNRS @agencerecherche.bsky.social

🥇 Félicitations à Lali Sakvarelidze-Achard, lauréate du #Photowalk de l' @iphc-strasbourg.bsky.social ! Sa photo «Télescope à micro-alignement : aux frontières de la physique nucléaire», avec 17 autres, représentera le
@cnrs.fr au Global Physics Photowalk 2025 🌍📸