The genetic architecture of local adaptation is historically contingent https://www.biorxiv.org/content/10.64898/2026.02.01.703099v1
03.02.2026 08:31 — 👍 3 🔁 4 💬 0 📌 1Alex Mackintosh
@thymelicus.bsky.social
Population genetics | Uppsala
@thymelicus.bsky.social
Population genetics | Uppsala
The genetic architecture of local adaptation is historically contingent https://www.biorxiv.org/content/10.64898/2026.02.01.703099v1
03.02.2026 08:31 — 👍 3 🔁 4 💬 0 📌 1
New paper out: “allopatric” Drosophila species aren’t so allopatric after all. We show that most currently allopatric species pairs probably overlapped in the past and exchanged genes at levels similar to sympatric pairs. @evolletters.bsky.social
doi.org/10.1093/evle... [1/6]

Hit me up if you'd like to apply to a Data-driven postdoc fellowship with me in Sweden. 2 year salary, excellent community. Deadline March 31. I have a project idea on reference bias vs pangenome, but keen on hearing ideas. Please share broadly.
www.scilifelab.se/data-driven/...

I'm excited to share that our preprint is now available on bioRxiv! Our study challenges the widespread use of molecular genetic diversity as a predictor adaptive potential, with important implications for how genetic data is used to inform conservation decisions.
www.biorxiv.org/content/10.6...

GHIST logo
Congratulations to the winners of the 2025 Genomic History Inference Strategies Tournament challenges!
Among the 9 challenges, we had five winners: @alwaysrong.bsky.social, @adaigle.bsky.social, @andrewhvaughn.bsky.social, @thymelicus.bsky.social, @rgollnisch.bsky.social

We are excited to share our recent work on the surprising robustness of Ancestral Recombination Graph (ARG) inference tools to computational phasing errors, now available on BioRxiv: biorxiv.org/content/10.1....
This work is co-advised by @yundeng.bsky.social and Rasmus Nielsen.
1/7
Very happy to announce that the next POPGROUP meeting will take place in Lille on 7-9 January !
Follow @popgroup2026.bsky.social for all updates.
📢 Please share widely ! 📢
You went to preschool with Naoyuki Takahata?!
15.09.2025 20:08 — 👍 3 🔁 0 💬 1 📌 0HiC. Not particularly cheap, but it is probably the only (non-trio) method where you can accurately phase SNPs across a whole chromosome.
27.08.2025 21:08 — 👍 0 🔁 0 💬 1 📌 0🚨New paper out in PNAS🚨
Soil eDNA reflects regionally dominant species rather than local composition of tropical tree communities 🧬🌿🌳🌺🧬
We asked about the spatial scale of biodiversity captured by samples of DNA from tropical soils #Luquillo #eDNA. A short 🧵....
www.pnas.org/doi/10.1073/...
A model-free method for genealogical inference without phasing and its application for topology weighting https://www.biorxiv.org/content/10.1101/2025.07.22.666161v1
26.07.2025 05:32 — 👍 4 🔁 5 💬 0 📌 0Our new work on estimation of background selection (BGS) is up on bioRxiv.
We investigated the problem using simulations and also developed a method for joint inference of BGS, demography and partial selfing from windowed site frequency spectra along the genome.
Feedback is welcome. #BGS
Our new work on estimation of background selection (BGS) is up on bioRxiv.
We investigated the problem using simulations and also developed a method for joint inference of BGS, demography and partial selfing from windowed site frequency spectra along the genome.
Feedback is welcome. #BGS
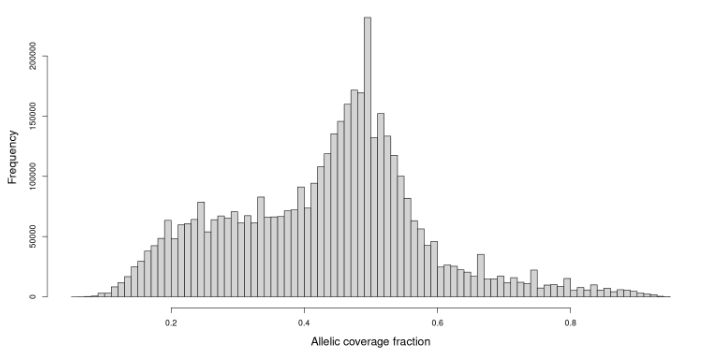
#PopGen New preprint of the team on the discovery of a very high rate of polymorphic duplication in a marine bivalve.
Imagine looking at the distribution of the allelic coverage fraction at heterozygous calls in an indivdual and getting this distribution 👇 😱😱😱
Speciation genomics in scarce swallowtails Iphiclides podalirius and I . feisthamelii : dx.plos.org/10.1371/jour...
28.04.2025 19:48 — 👍 11 🔁 5 💬 0 📌 0The 229 chromosomes of the Atlas blue butterfly reveal rules constraining chromosome evolution in Lepidoptera https://www.biorxiv.org/content/10.1101/2025.03.14.642909v1
17.03.2025 19:32 — 👍 15 🔁 5 💬 0 📌 1Likelihoods for a general class of ARGs under the SMC https://www.biorxiv.org/content/10.1101/2025.02.24.639977v1
28.02.2025 03:33 — 👍 11 🔁 8 💬 0 📌 0
https://coursesandconferences.wellcomeconnectingscience.org/event/k-mer-workshop-for-biodiversity-genomics-20250601/
🚨Registration open🚨 1st - 6th of June, me, Katie Jenike, @rayanchikhi.bsky.social and Gene Myers will run a week long workshop on k-mers for biodiversity genomics organised by @connectingscience.bsky.social at @sangerinstitute.bsky.social. Join us & get equipped to wrestle large genomic datasets!
31.01.2025 15:17 — 👍 26 🔁 17 💬 1 📌 1The test requires just one unphased diploid from each population and makes few assumptions about the underlying demographic history. I hope that others find the paper/method useful. 2/2
12.12.2024 14:17 — 👍 6 🔁 2 💬 1 📌 0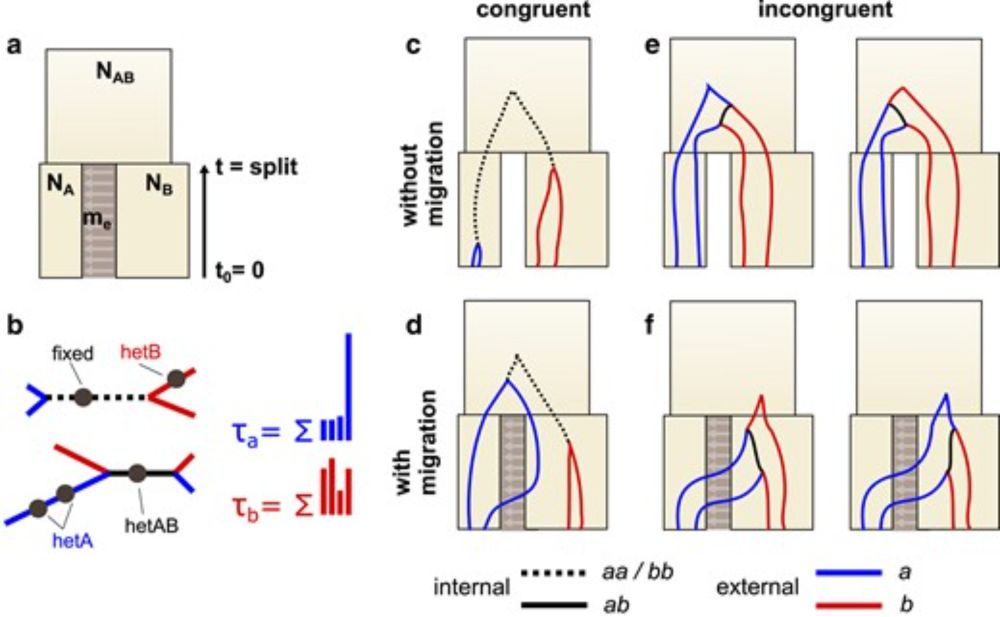
My and Derek Setter's work on gene flow is now published in the latest issue of Genetics. We show that it is possible to detect past gene flow between two populations by summarising the asymmetry in pop-specific external branch lengths. 1/2
tinyurl.com/5ff8ch9e
Sadly I am no longer working on butterfly genomics. I am currently focused on popgen methods with application to plant genomes (mostly simulated plant genomes for now...) Perhaps I'll return one day 🦋
03.12.2024 14:14 — 👍 3 🔁 0 💬 0 📌 0
Aricia species photos
A new case of hybrid speciation? www.biorxiv.org/content/10.1...
20.11.2024 15:41 — 👍 13 🔁 6 💬 1 📌 0