At #EASD and interested in learning about how polygenic risk scores can be use to improve prediction and prevention of type 2 diabetes? Come see my talk at 17:25 today in the Milan Hall as part of the EASD-ADA Joint Symposium. I'll also be around until Friday afternoon if anyone wants to chat.
16.09.2025 12:44 — 👍 2 🔁 0 💬 0 📌 0
Research Associate*/Research Assistant in Sustainability of AI (Fixed Term)
An exciting opportunity has arisen for a talented researcher to join our team as part of the Green Algorithms Initiative, one of the leading academic teams in the field of sustainable computing. The
⏰ Last couple of days to apply to join my group @Cambridge as a postdoc and work on the environmental (un)sustainability of AI!
⏳ Closing September 16th.
✏️ Apply here: tinyurl.com/2ukkp8yx
Or learn more about what we do at www.lannelongue-group.org
Initial examples of research projects below 👇
15.09.2025 08:50 — 👍 10 🔁 10 💬 1 📌 0
prcomp() uses BLAS, which depending on the BLAS library and how R was compiled against it when R was installed will default to using all available cores.
Try adding the following code before your call to prcomp():
library(RhpcBLASctl)
blas_set_num_threads(1)
omp_set_num_threads(1)
15.08.2025 11:58 — 👍 7 🔁 0 💬 1 📌 0

Reminds me of this classic tweet:
07.08.2025 22:14 — 👍 1 🔁 0 💬 0 📌 0
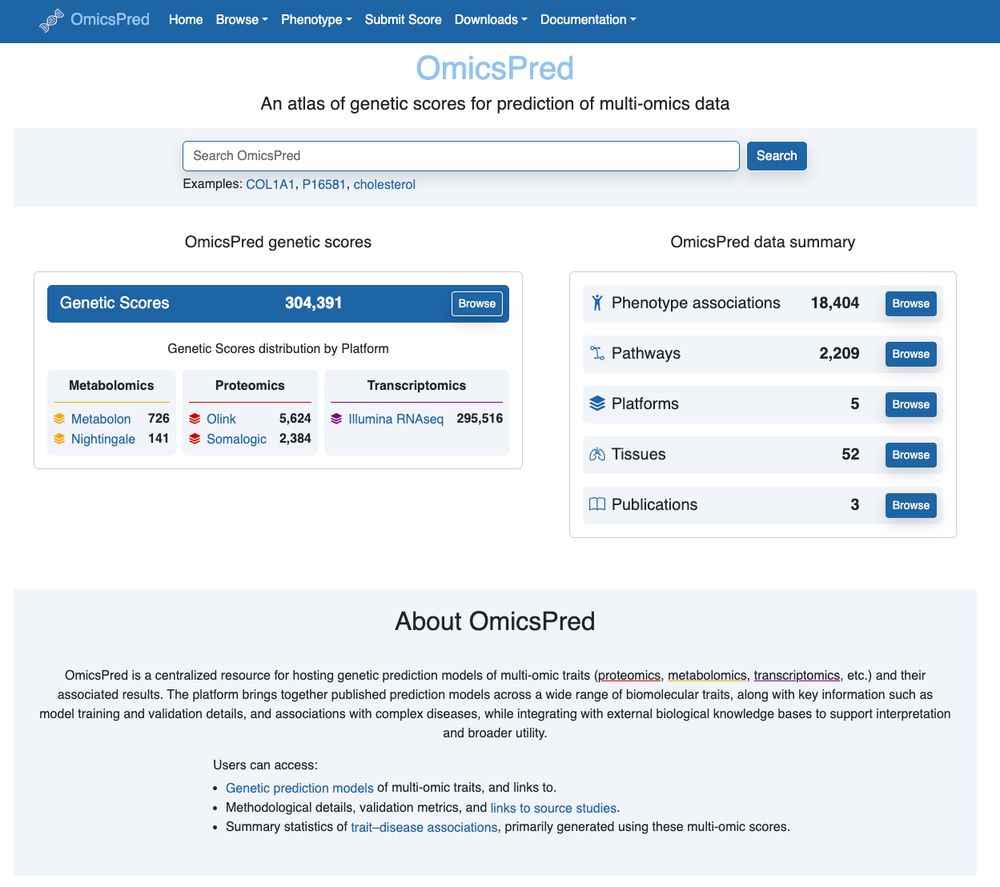
Super excited to see beginning integration of PredictDB into OmicsPred!
We've annotated/deposited the GTExV8 gene expression predictors so they are now available alongside all the other multi-omic predictors at www.omicspred.org
More on its way... a wonderful collab with @hakyim.bsky.social & co!
16.06.2025 10:33 — 👍 19 🔁 8 💬 0 📌 0
Has Antony Green called the Eurovision winner yet? #eurovision #auspol #stillwaitingfortheprepolls
17.05.2025 22:29 — 👍 3 🔁 1 💬 1 📌 0
Estonia hands down
17.05.2025 21:31 — 👍 5 🔁 2 💬 1 📌 0
Feeling mildly emotional that this is Green’s last election coverage. He’s covered every Aus election I’ve ever seen, it won’t be the same without him #auspol
03.05.2025 06:56 — 👍 15 🔁 1 💬 1 📌 0

A screenshot of a search for the NCBI gene website on the internet wayback machine showing snapshot archives available between 2008 and January 2025
Now's a good time for a reminder about the internet wayback machine: web.archive.org
06.02.2025 16:40 — 👍 0 🔁 0 💬 0 📌 0
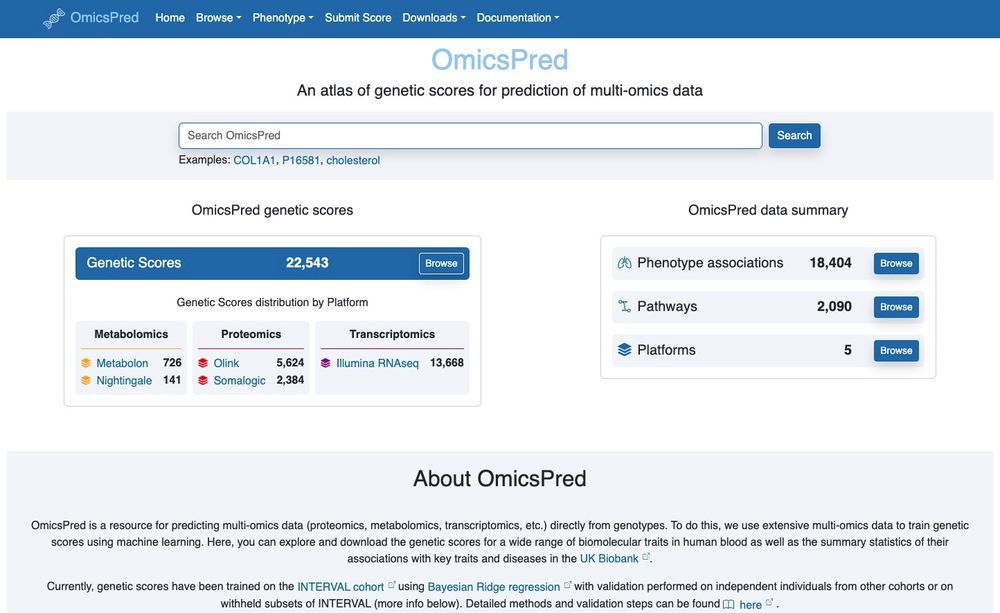
Introducing a major upgrade to OmicsPred platform (www.omicspred.org) — a resource to enhance the accessibility and usability of genetic scores for multi-omic traits and their phenotypic associations. (1/N)
06.02.2025 11:09 — 👍 15 🔁 13 💬 1 📌 2

A platform for the biomedical application of large language models https://www.nature.com/articles/s41587-024-02534-3 (read free: https://rdcu.be/d68Ih) MIT licensed https://github.com/biocypher/biochatter 🧬🖥️🧪
22.01.2025 16:30 — 👍 11 🔁 1 💬 0 📌 0

Flyer for the LPSHG - details in the post
Excited to be tutoring at the Leena Peltonen School of Human Genetics on July 27-31, alongside a stellar crew. If you’re a late-stage or recently graduated PhD student this is an awesome opportunity to get 1:1 time with faculty at the cutting edge of genomics.
Apply by March 7th at lpshg.com
19.12.2024 11:37 — 👍 10 🔁 3 💬 0 📌 0
at first glance this just looks like ggplot but 's/geom/add/' 🤔
21.11.2024 20:04 — 👍 1 🔁 0 💬 0 📌 0
⏳ Just one more week to apply! 5 roles open in my group working on the Green Algorithms project: 3 research, a Software Engineer Lead and a Community manager/project coordinator
🌱 More details and links to all adverts there: www.green-algorithms.org/join-us/
Closing 25/11
#AcademicSky #SciSky
19.11.2024 10:08 — 👍 9 🔁 5 💬 0 📌 0

Figure 4 from the preprint
Modelling the impact of these in the wider UK population, we show that supplementing screening with conventional risk factors (i.e. SCORE2) with targeted follow-up with NMR scores and PRSs could increase the # of CVD events prevented from 201 to 370 per 100K screened
Thread 3/3
02.11.2023 13:05 — 👍 1 🔁 0 💬 0 📌 0

Figure 3 from the preprint
Using the latest UK Biobank data, we show that:
- Integrating NMR scores and PRSs with SCORE2 can moderately improve 10-year CVD risk prediction (by up to 9%)
- Risk stratification is also improved when using ESC 2021 guideline-recommended risk categorisation
Thread 2/3
02.11.2023 13:04 — 👍 1 🔁 0 💬 1 📌 0
For my debut to bluesky, I am pleased to announce a new preprint:
medrxiv.org/content/10.1...
Here, we show that NMR biomarker scores combined with PRSs and SCORE2 may have moderate population health benefits for 10-year CVD risk prediction prevention
Thread 1/3:
02.11.2023 13:02 — 👍 7 🔁 4 💬 1 📌 0
COMBINE is a national student society for students in computational biology, bioinformatics, and related fields. COMBINE is the student subcommittee of ABACBS as well as the official ISCB regional student group for Australia.
Proteomics, PTMs and molecular epidemiology
Royal Commission for the Exhibition of 1851 Research Fellow
Emmanuel College Research Fellow
University of Cambridge
Human genetics, type 2 diabetes and obesity in today's environment. Cycling uphill. Team science. New shoots in Geneva & France, long roots in the UK & Exeter. https://www.unige.ch/medecine/gede/en/research-groups/timothy-frayling
statistical geneticist; postdoc in the Singh Lab at NYGC; previously BU biostat and Carleton College
The CEU is an internationally-renowned centre of excellence for the study of the causes and prevention of cardiovascular disease.
https://www.phpc.cam.ac.uk/research/departmental-research-units/cardiovascular-epidemiology-unit-ceu
Australian Election Analyst
Weird and wild moments from Down Under. Smash follow!
(And for clarity, just because something's Batshit that doesn't mean we think it's bad)
BHF Chair of Cardiovascular AI & Consultant Radiologist. Computational Cardiac Imaging group, MRC Laboratory of Medical Sciences. 🚁
Group Leader
Computational Biology
Peter MacCallum Cancer Centre
Follow for research, news, events and other stories from the Department of Public Health and Primary Care.
Strategy & Partnering @Metabolon
Multiomics | Science | Outdoor life
#metabolomics
https://www.linkedin.com/in/timdelany/
Genetic epidemiology and pharmacogenomics research, U de Montréal
Online-only Nature Research journal that publishes basic, translational, clinical and public health research in cardiac, vascular and blood biology.
https://www.nature.com/natcardiovascres/
A resource to enhance the accessibility and usability of genetic scores for multi-omic traits and their phenotypic associations
www.omicspred.org
Passionate about #GWAS #Metabolomics #Proteomics #Glycomics #Epigenomics, Professor @WeillCornell
Medicine - Qatar, Blog on http://metabolomix.com, http://suhre.fr
Building sustainable healthcare
Learn more ➡️ linktr.ee/nightingalehealth
BHF PhD student working on CVD risk communication and health equity at Cambridge Uni (they/he) 🌈📊
Public Health | Epidemiology | Nutrition | Health Risk Communication | Health Equity | Prevention
https://cosoc.com/OwenATaylor
Senior Research Fellow at Menzies Institute for Medical Research
Molecular evolution, human genetics, bioinformatics