Congrats @hoheyn.bsky.social , @lgmartelotto.bsky.social and team!
23.06.2025 07:13 — 👍 4 🔁 2 💬 0 📌 0

Counting my lucky stars⭐️that not only do I get to work with these amazing scientists Jiwoon Park @christophermason.bsky.social but also give fun @cshlnews.bsky.social lectures together💪.Big thank you to @lgmartelotto.bsky.social Nicholas Navin Julea Vlassakis for uniting this #singlecell #dreamteam
17.07.2025 15:46 — 👍 5 🔁 2 💬 0 📌 0
Seminar Series – GESTALT – Global Alliance for Spatial Technologies
Don’t forget to register to the seminar on June 27 and July 25:
GESTALT - Seminar Series
June 27 - 2025: Dr. Fei Chen
July 25 - 2025: Dr. Hanchen Wang
Please visit the Seminar Series Webpage for more information and registration: globalspatial.org/seminar-seri...
22.06.2025 22:35 — 👍 2 🔁 2 💬 0 📌 0
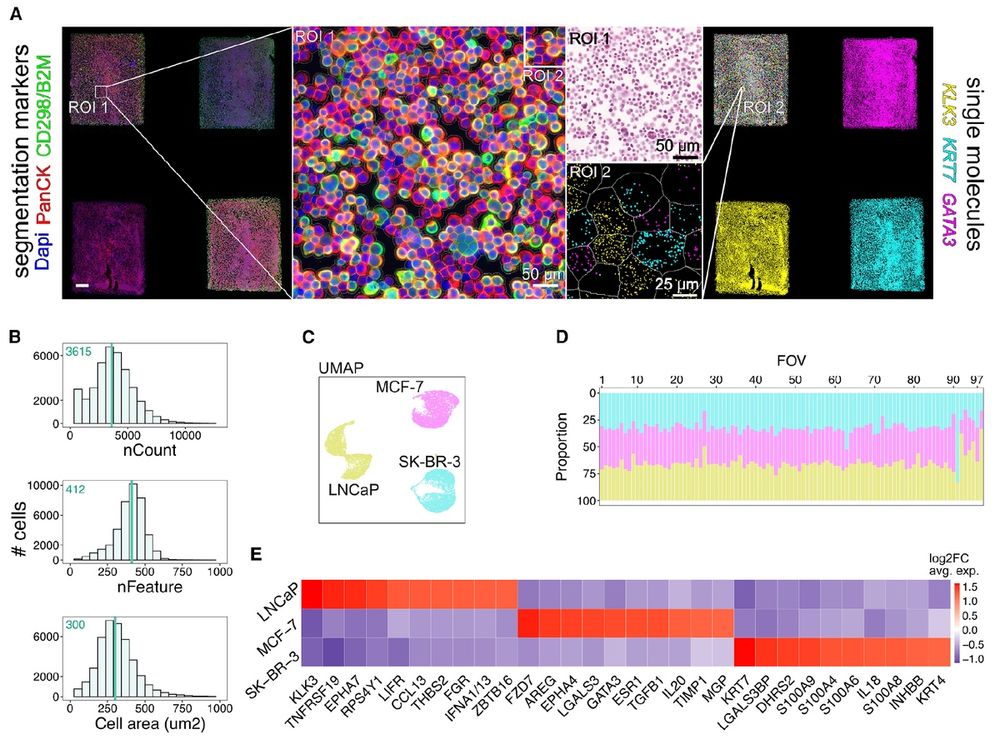
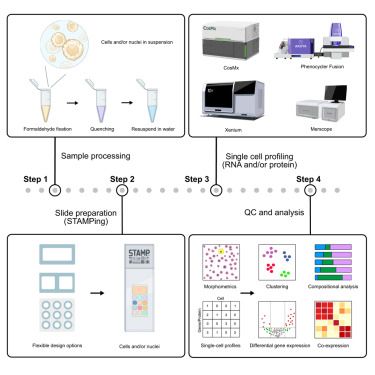
STAMP:Single cell Transcriptomics Analysis & Multimodal Profiling-cheap scprofiling-MILLIONS of cells, multimodal, pertubation expts & tissue atlas'ing at scale.Thx to brilliant colleagues @lgmartelotto.bsky.social Holger Heyn & our amazing teams. @cellpress.bsky.social www.cell.com/cell/abstrac...
17.06.2025 21:47 — 👍 12 🔁 4 💬 0 📌 0
“STAMP allows us to profile a million cells simultaneously, compared to tens of thousands typical of current methods, making it far more scalable,” said @jasmineplummer.bsky.social, PhD, St. Jude Center for Spatial Omics director.
17.06.2025 15:01 — 👍 4 🔁 3 💬 0 📌 1
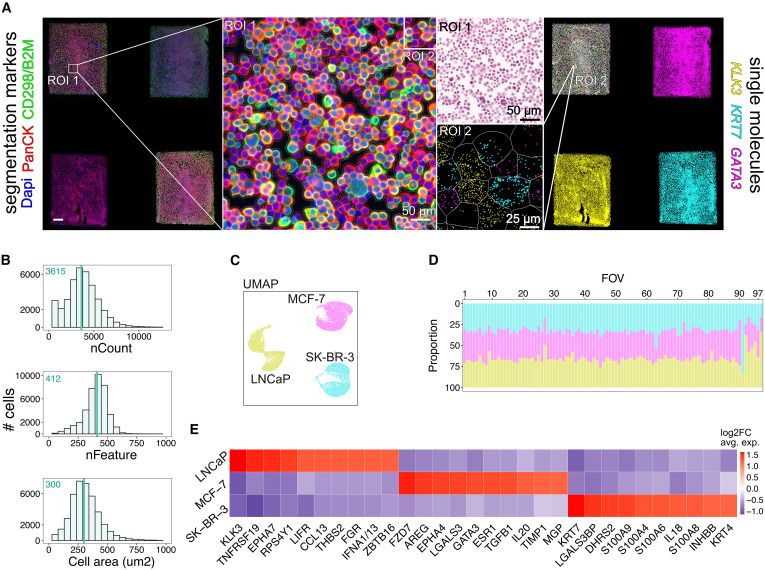
STAMP: Single-cell transcriptomics analysis and multimodal profiling through imaging @cellcellpress.bsky.social @lgmartelotto.bsky.social @hoheyn.bsky.social
www.cell.com/cell/fulltex...
17.06.2025 16:40 — 👍 19 🔁 11 💬 0 📌 1

👩🔬The lead authors of the study are Dr. @pascual-reguant.bsky.social & Emanuele Pitino (CNAG) with Felipe Segato Dezem (@stjuderesearch.bsky.social)
Corresponding authors: Dr. @hoheyn.bsky.social (CNAG), Dr. @jasmineplummer.bsky.social (St. Jude) & Dr. @lgmartelotto.bsky.social (Univ. of Adelaide)
17.06.2025 15:39 — 👍 5 🔁 3 💬 0 📌 0
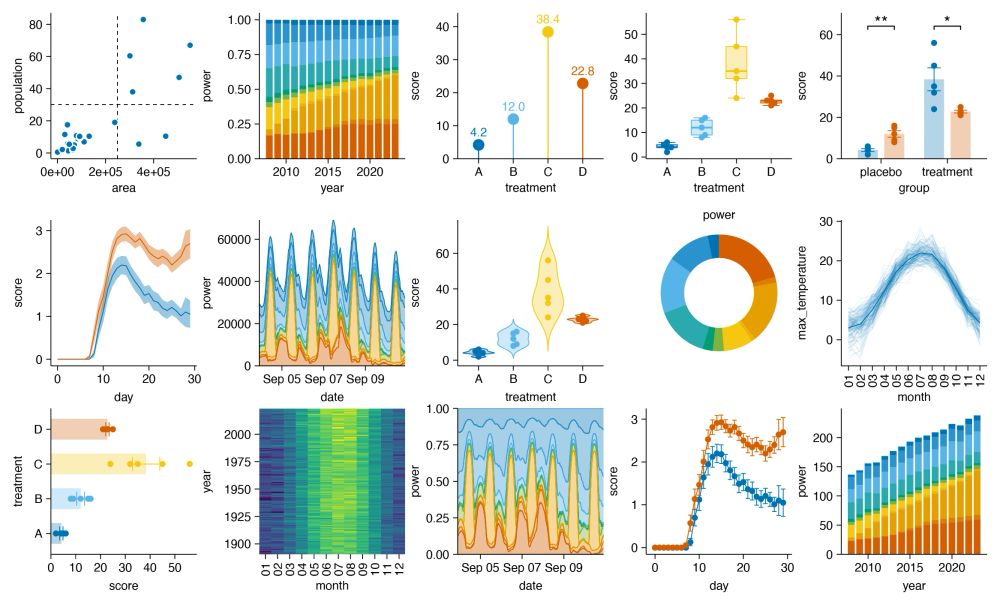
tidyplots is an R package to generate publication-ready plots for scientific papers tidyplots.org/ #rstats
25.12.2024 14:45 — 👍 92 🔁 15 💬 1 📌 3
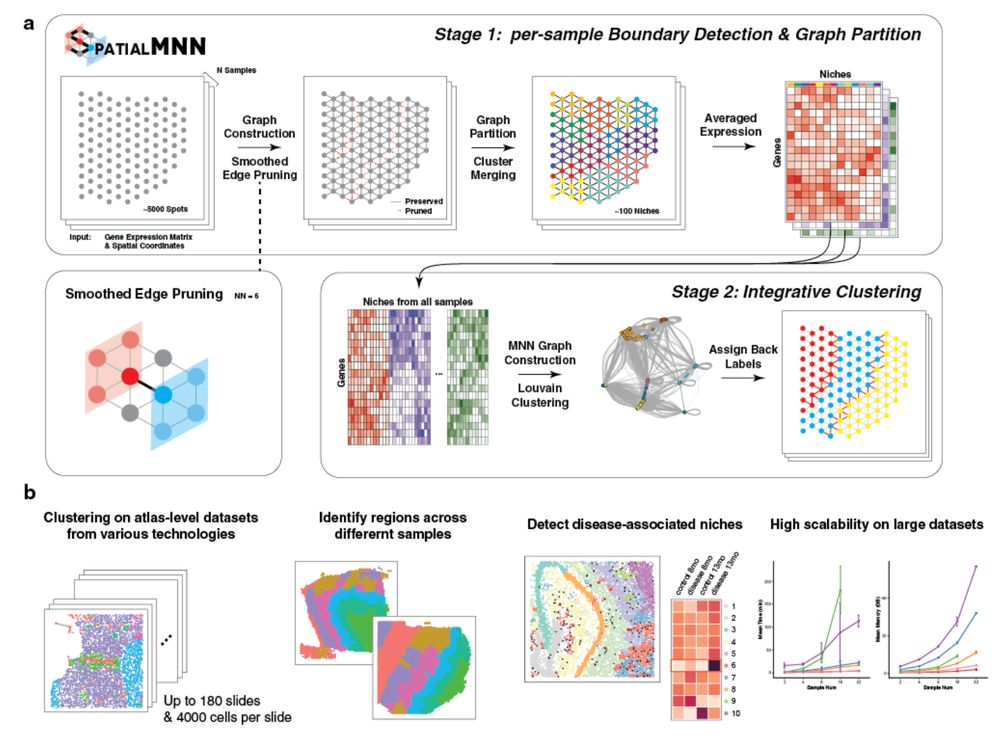
Excited to share a new algorithm that we have been working on over the last year.
💡 idea is to extend mutual nearest neighbors for
#spatial data. We call it spatial mutual nearest neighbors (spatialMNN) 😄
Thank you @haowen-zhou.bsky.social @pratibha-panwar.bsky.social who led this work! 👏 🧬🖥️🧪
05.12.2024 16:31 — 👍 93 🔁 27 💬 4 📌 1

It is so nice to spend quality time with your favorite people in the world before the end of the year!
@jasmineplummer.bsky.social @alexswarbrick.bsky.social
At Multi-Ome 2024
Btw: my best pic yet!
03.12.2024 14:18 — 👍 4 🔁 1 💬 0 📌 0
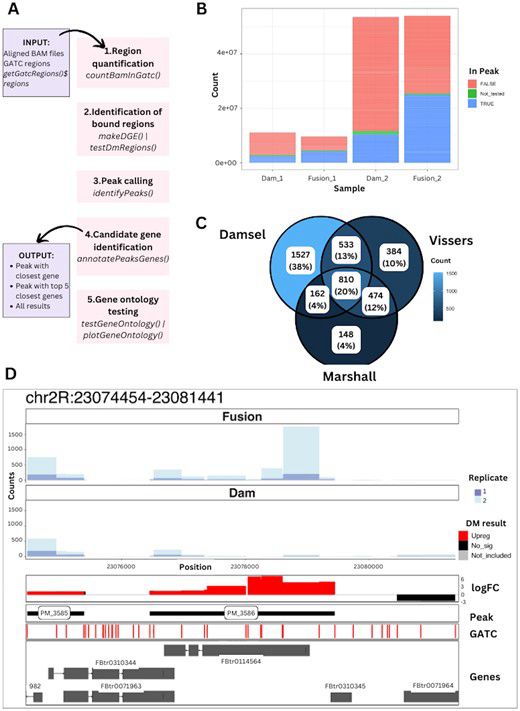
Damsel: analysis and visualisation of DamID sequencing in R
AbstractSummary. DamID sequencing is a technique to map the genome-wide interaction of a protein with DNA. Damsel is the first Bioconductor package to prov
Happy to see our Damsel @bioconductor.bsky.social package published in Bioinformatics. Great work putting together this DamID sequencing and package from student Caitlin Page. Thanks to collaborators too @lonsbio.bsky.social
academic.oup.com/bioinformati...
29.11.2024 02:11 — 👍 23 🔁 7 💬 0 📌 1
It is important to clarify what you mean by real science. If ‘real’ refers to methodologically sound, evidence-driven, and reproducible research, then funding may not directly define the quality of the science, but it certainly influences the scale, tools, and opportunities available for doing it.
30.11.2024 17:42 — 👍 0 🔁 0 💬 0 📌 0
Whole-brain spatial transcriptomics 🧪🤯
Congratulations to all the team @peruhlen.bsky.social
24.11.2024 10:49 — 👍 29 🔁 4 💬 0 📌 1
Cellular atlases are unlocking the mysteries of the human body
Scientists reflect on insights from the Human Cell Atlas.
The Human Cell Atlas is yielding detailed maps of human tissues and systems throughout life, along with methods to handle single-cell data. Four scientists reflect in Nature on how the project is transforming our understanding of human biology. 🧪
26.11.2024 15:06 — 👍 131 🔁 32 💬 0 📌 0
GitHub - theislab/spapros: Python package for Probe set selection for targeted spatial transcriptomics.
Python package for Probe set selection for targeted spatial transcriptomics. - theislab/spapros
#LocationLocationLocation! Theis &co share @ Nature Methods @natureportfolio.bsky.social a probe set selection pipeline to identify cell types using spatial single cell omics! And it comes with its Python package (github.com/theislab/spa...) 😌💪 👉 doi.org/10.1038/s41592-024-02496-z
20.11.2024 12:30 — 👍 8 🔁 1 💬 0 📌 0
 03.10.2025 23:54 — 👍 3 🔁 2 💬 0 📌 0
03.10.2025 23:54 — 👍 3 🔁 2 💬 0 📌 0