
Happy to share our latest publication, in which we show that the arrangement of nucleosomes around CTCF sites contributes to higher-order organisation of chromatin into TADs: www.embopress.org/doi/full/10....
27.10.2025 11:49 — 👍 71 🔁 31 💬 0 📌 2@kasitc.bsky.social
Postdoctoral Fellow, Genome Biology Unit, EMBL Heidelberg

Happy to share our latest publication, in which we show that the arrangement of nucleosomes around CTCF sites contributes to higher-order organisation of chromatin into TADs: www.embopress.org/doi/full/10....
27.10.2025 11:49 — 👍 71 🔁 31 💬 0 📌 2Out now! 🎉 Check the thread & preprint to see why we think E–P specificity is real in mammals — and, well, a few other interesting things popped up too 👀
Huge thanks to @danielibrahim.bsky.social, @arnaudkr.bsky.social & @stemundi.bsky.social and fantastic people in their labs — what a journey! 🧪🔬
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
Have you ever wondered how the exact location of a gene affects it's activity?
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you

ikea-style logo of splongget
1/ First preprint from @jdemeul.bsky.social lab 🥳! We present our new multi-modal single-cell long-read method SPLONGGET (Single-cell Profiling of LONG-read Genome, Epigenome, and Transcriptome)! www.biorxiv.org/content/10.1...
10.09.2025 15:48 — 👍 48 🔁 16 💬 1 📌 1
Fragile Nucleosome Fall Seminars: September 10 Grace Bower, Kvon Lab @ UC Irvine, USA Jennifer Phillips-Cremins, UPenn, USA September 24 Ali Wilkening, Sanulli Lab @ Stanford, USA Juanma Schvartzman, Columbia University, USA October 8 Sanim Rahman, Greenberg Lab @ UPenn, USA Alex Federation, Talus Bio, USA October 22 Alice Laigle, Croll Lab @ University of Neuchâtel, Switzerland Seungsoo Kim, UC Irvine, USA November 5 Hannah Long, University of Edinburgh, UK Jeff Vierstra, Altius Institute, USA November 19 Ishtiaque Hossain, Pastor Lab @ McGill, Canada Sarah Teichmann, University of Cambridge, UK December 3 María Mariner Faulí, Rada Iglesias Lab @ IBBTEC, Spain Jonathan Henninger, Carnegie Mellon, USA December 17 Rebecca Berrens, Oxford University, UK Jean-Benoit Lalanne, University of Montreal, Canada
We're super excited to announce the entire lineup for the Fall season of Fragile Nucleosome Seminars, starting on Sept 10th at 1200 EDT / 1600 UTC with @gracebower.bsky.social and @creminslab.bsky.social!
register here for the entire series: us06web.zoom.us/webinar/regi...

Excited / nervous to share the “magnum opus” of my postdoc in Andreas Wagner’s lab!
"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)
Congrats Pedro, glad to see you write about ageing & cancer again!
26.08.2025 20:35 — 👍 1 🔁 0 💬 0 📌 0
New paper @natrevcancer.nature.com on the evolutionary interplay between ageing and cancer 🚨
www.nature.com/articles/s41...
Please RP.
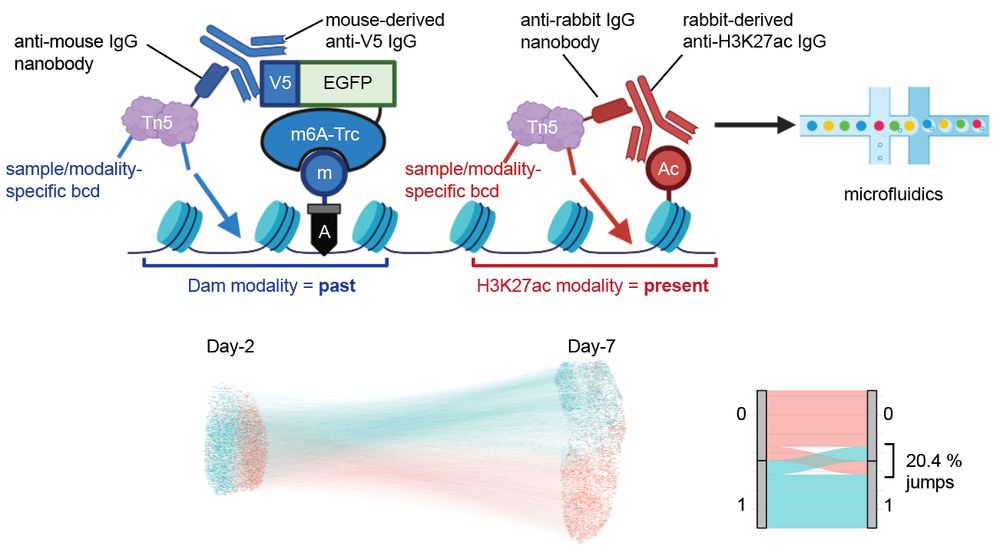
We are thrilled to announce that our lab’s first preprint is out!
”Whole-genome single-cell multimodal history tracing to reveal cell identity transition”
We report HisTrac-seq, a multiomic single-cell molecular recording platform.
www.biorxiv.org/content/10.1...

Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation www.biorxiv.org/content/10.1... @embl.org
18.08.2025 12:39 — 👍 19 🔁 4 💬 2 📌 0Huge thanks to the co-first author @mathias-boulanger.bsky.social. Also thanks to Rene, Marc Jan, Karine, Kim, and especially @arnaudkr.bsky.social @embl.org @dfg.de.
Finally, I'll be presenting the story at the @cshlnews.bsky.social #cshlmoet next week! Please stop by if you are around!

What about transcription? We could link changes in enhancer activity to the consequences at a co-accessible promoter!
18.08.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Can we prove coordination in their activity? Using regulatory changes between cell types, we show that co-accessible enhancers co-vary, demonstrating the dependency between them.
18.08.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
What regulates co-accessibility? We found that certain TF motifs are enriched in co-accessible CREs (vs independent ones). These include several usual suspects such as Su(Hw) and Trl (GAF).
18.08.2025 12:23 — 👍 2 🔁 0 💬 1 📌 0
Co-accessible pairs aren't always the closest neighbours! For each pair of CREs, we tested if they are co-accessible more often than expected by chance, creating a co-accessibility map across CREs.
18.08.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Here is an example locus: we can resolve chromatin accessibility for a promoter and 8 enhancers located up to 20kb away! — all on the same DNA molecules.
18.08.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
We used Single-molecule footprinting (SMF) to methylate accessible regions, combined with Nanopore long-read sequencing. At each locus, we collected thousands of molecules — enough to quantify coordinated accessibility across distant cis-regulatory elements (CREs) up to 30 kbs apart!
18.08.2025 12:23 — 👍 0 🔁 0 💬 1 📌 0
Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation. www.biorxiv.org/content/10.1...
18.08.2025 12:23 — 👍 97 🔁 39 💬 2 📌 2
Excited to see this published with additional data following our preprint a while back. Cool combination (in our biased view) of controlled TF expression and machine learning to decode chromatin sensitivity. www.sciencedirect.com/science/arti....
07.08.2025 16:20 — 👍 114 🔁 49 💬 2 📌 3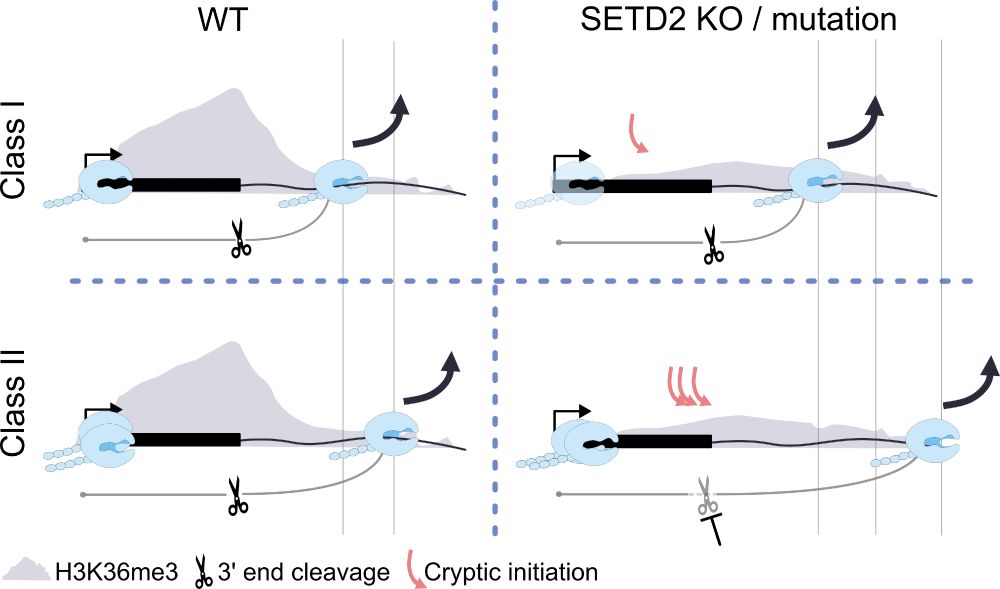
🚨 New preprint from our labs (STOP lab @stoplab.bsky.social & Nojima lab @pol2rna.bsky.social @pyrolyn.bsky.social)! We've been digging into how chromatin remodeler SETD2 controls the start and end of transcription 👇 www.biorxiv.org/cgi/content/... (1/5)
16.07.2025 09:38 — 👍 12 🔁 6 💬 6 📌 0
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
02.07.2025 16:17 — 👍 186 🔁 90 💬 10 📌 9
Join us next Wed, for three amazing ECR researchers_ @fueyoraquel.bsky.social, @kaiamattioli.bsky.social and Laura talk about their work in #FragileNucleosome seminar series!
In case you haven't joined any of the previous 2025 sessions, here is registration link:
us06web.zoom.us/webinar/regi...

How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
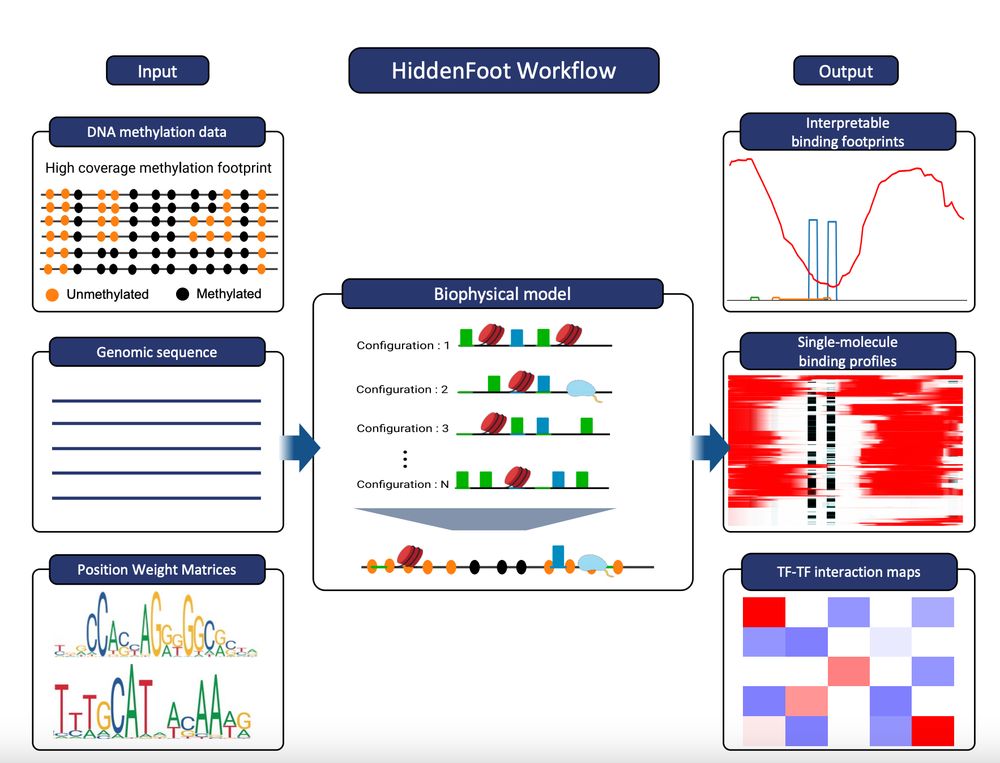
🚨 New preprint out! Do you think Single Molecule Footprinting and Fiber-seq are super cool but aren't sure how to unlock their full potential? HiddenFoot can help you: www.biorxiv.org/content/10.1...
17.05.2025 17:00 — 👍 20 🔁 5 💬 0 📌 0
Join us on Wednesday next week for two exciting talks on transcription regulation from @kasitc.bsky.social and @davidsuter.bsky.social!
You can register at: us06web.zoom.us/webinar/regi...

5/2025 Issue ➡️ www.embopress.org/toc/17444292...
tissue-specific huntingtin interactomes, gene product diversity evolution
Cover: mammalian promoters characterised by low RNA pol II occupancy and high turnover @kasitc.bsky.social @molinalab.bsky.social @arnaudkr.bsky.social
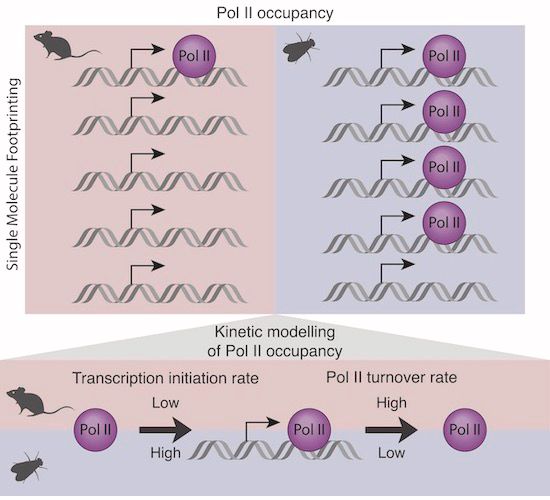
New research by @kasitc.bsky.social @molinalab.bsky.social @arnaudkr.bsky.social reveals limited RNA polymerase II pausing at mouse promoters compared to flies ➡️ www.embopress.org/doi/full/10....
08.05.2025 15:19 — 👍 5 🔁 6 💬 0 📌 0

Pol II Pausing on LSD on the MSB cover! Credits to the amazing creative team @kasitc.bsky.social (right) and Roos (left)! Pleasure to work with this bunch of talented people, here at our Belgian retreat. Paper: doi.org/10.1038/s443...
02.05.2025 09:30 — 👍 23 🔁 6 💬 1 📌 0Don't forget to checkout our annex pre-print bit.ly/3EkRqJh for a sense on our new 💻 tool FootprintCharter, for the unsupervised footprint quantification from single molecule data. Available on Bioconductor at bit.ly/3XLe8RC.
08.04.2025 15:52 — 👍 7 🔁 3 💬 0 📌 0