
Schopenhauer
11.01.2026 01:11 — 👍 222 🔁 29 💬 5 📌 1@tkorem.bsky.social
Microbiome, metagenomics, ML, and reproductive health. All views are mine. So are all your base

Schopenhauer
11.01.2026 01:11 — 👍 222 🔁 29 💬 5 📌 1Pollution dropping by that much likely means a whole bunch of people are alive who otherwise wouldn’t be. Incredible stuff.
23.12.2025 22:00 — 👍 105 🔁 21 💬 1 📌 0
We just published in @molsystbiol.org with the Mugler lab (UPitt) on bacterial population dynamics during tumor colonization (mouse model). Our study was guided by a Luria–Delbrück-style idea: infer mechanism from statistics (1/7) 🧪🦠
doi.org/10.1038/s443...
Out after peer-review: www.science.org/doi/full/10....
Our bottom line stayed: never use leave-one-out cross-validation as it has inherent train-test leakage. Consider our Rebalanced version instead!
We now also account for regression and nested cross-validation, with more extensive benchmarking.
Using leave-one-out cross-validation to calculate metrics such as AUC and R^2 creates bias! This can be fixed by removing one of each class in the meanwhile to maintain the training data distribution - great work by Tal and his team 🧪🧬🖥️
29.11.2025 13:04 — 👍 11 🔁 3 💬 0 📌 0
I found a flowchart which helps you navigate the IT landscape
01.10.2025 18:22 — 👍 10584 🔁 3748 💬 53 📌 65Back when we hid beer in the cold room in a box labeled “yeast embryos.”
29.11.2025 00:07 — 👍 14 🔁 2 💬 1 📌 0Out after peer-review: www.science.org/doi/full/10....
Our bottom line stayed: never use leave-one-out cross-validation as it has inherent train-test leakage. Consider our Rebalanced version instead!
We now also account for regression and nested cross-validation, with more extensive benchmarking.
As long as three reviewers keep reading each proposal, doesn't really address anything either
13.11.2025 23:23 — 👍 1 🔁 0 💬 0 📌 0"This could take down Democrats, too."
I know. And I frankly wouldn't give even an itty bitty damn if it implicated every Democratic man in Congress, every Democratic hopeful for 2028 and every Democrat who has even thought about running for office.
Down with the sex predators, wherever they are.
White paper: github.com/aqlaboratory...
GitHub Repo: github.com/aqlaboratory...
Huggingface: huggingface.co/OpenFold/Ope...
Testimonials: openfold.ghost.io/openfold3-co...
OpenFold3-preview (OF3p) is out: a sneak peek of our AF3-based structure prediction model. Our aim for OF3 is full AF3-parity for every modality. We now believe we have a clear path towards this goal and are releasing OF3p to enable building in the OF3 ecosystem. More👇
28.10.2025 18:30 — 👍 125 🔁 42 💬 1 📌 3Just to be super clear, if you’re phoning in your peer review to ai you should quit your job so someone else who actually likes science can have it.
16.10.2025 23:37 — 👍 371 🔁 67 💬 6 📌 2No idea. Still working through this.
19.09.2025 03:06 — 👍 1 🔁 0 💬 0 📌 0This is from the very paper you linked to - Figure S5. They claim that this has a p-value of 1. It's not here and there, this is what most results look like, and this is a large part of the basis for claiming that there are no robust associations with other tumors.
19.09.2025 03:01 — 👍 0 🔁 0 💬 1 📌 0
So you look at this figure and your interpretation is "no signal"?
18.09.2025 23:50 — 👍 1 🔁 0 💬 1 📌 0I never knew I needed this thread
16.08.2025 02:48 — 👍 1 🔁 0 💬 0 📌 0
We are hiring a postdoc - come work with us (www.koremlab.science) at the intersection of #microbiome, data science, and women's health!
Message or email me if interested. 🖥️ 🧬
I am seeking a postdoc for my group at UCLA. We work at the intersection of population genetics x microbiome (garud.eeb.ucla.edu). If interested, please message me!
22.07.2025 17:51 — 👍 85 🔁 100 💬 1 📌 4This will also likely reduce the number of study sections, firing SROs and making them less specialized.
17.07.2025 21:51 — 👍 0 🔁 0 💬 1 📌 0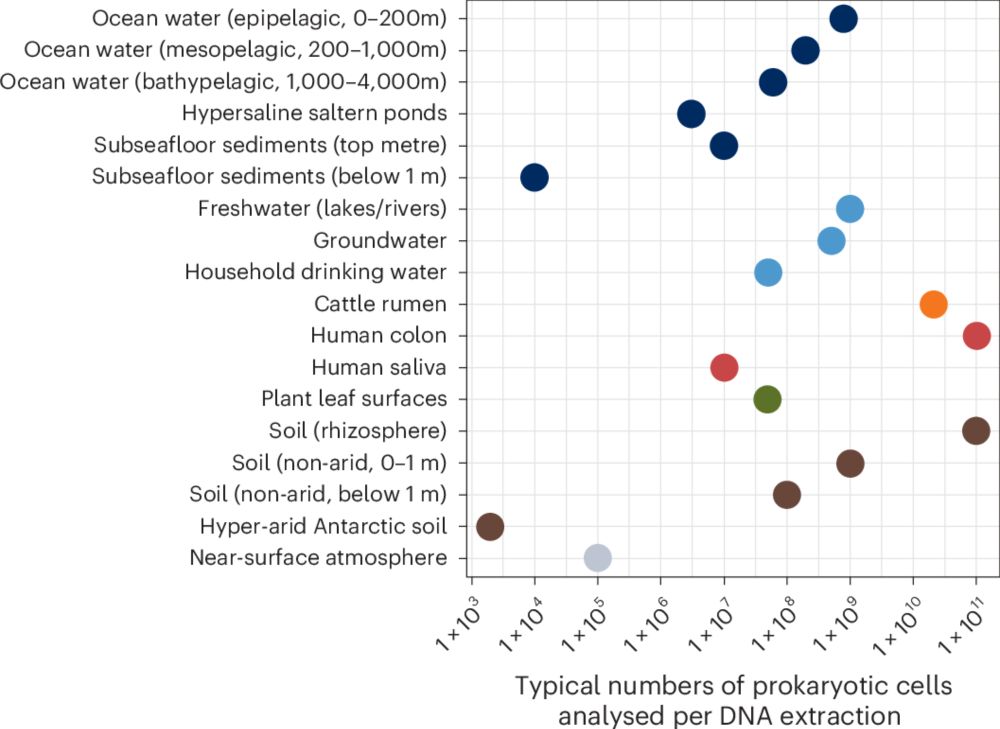
Hope this is useful - consensus statement "Guidelines for preventing and reporting contamination in low-biomass microbiome studies" rdcu.be/er3Io
21.06.2025 19:59 — 👍 106 🔁 48 💬 2 📌 2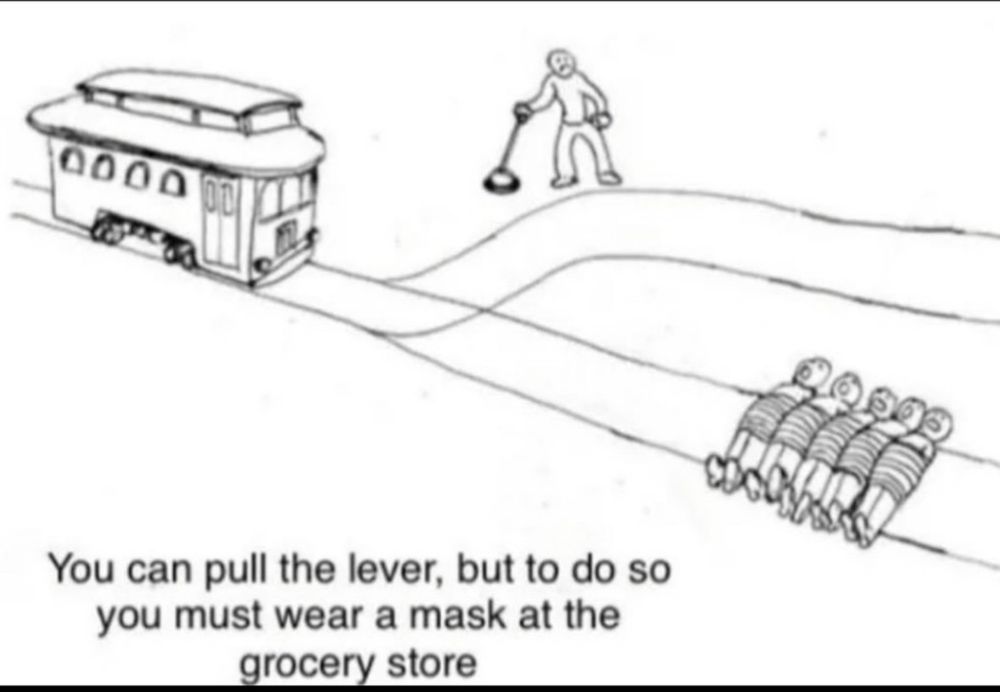
The trolley problem graphic where one side is several people tied to the train tracks except the other side has no one on the tracks. A man is standing holding a lever trying to decide which track the trolley should take. The text reads “You can pull the lever, but to do so you must wear a mask at the grocery store.”
25.05.2025 20:55 — 👍 1487 🔁 433 💬 7 📌 13
Not getting much attention, except in a recent NYTimes story, is a provision to increase the tax on university endowments and those of other nonprofits.
22.05.2025 10:22 — 👍 736 🔁 373 💬 27 📌 33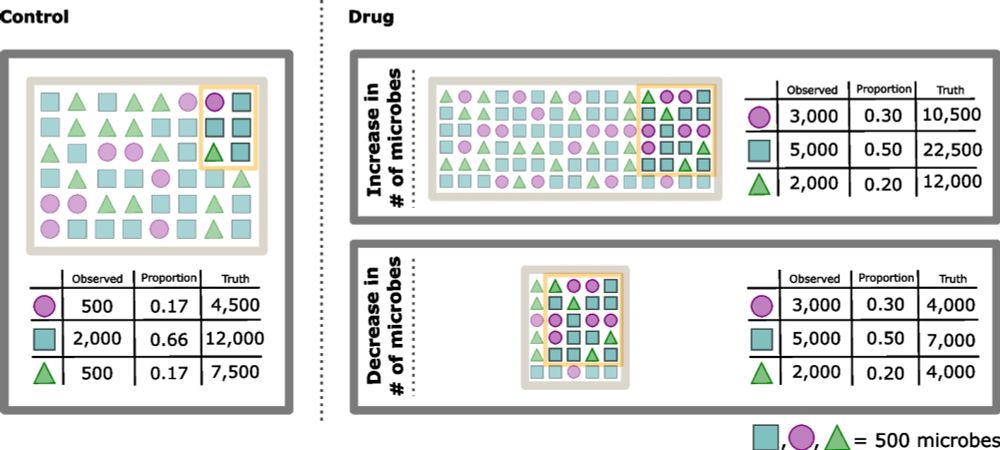
New paper in Genome Biology!
genomebiology.biomedcentral.com/articles/10....
We introduce scale models, a generalization of normalizations that explciitly account for uncertainty in biological system scale (e.g., microbial load).
as a resident of syracuse, ny, a rust belt town that used to be an economic epicenter for the nation: syracuse university is our largest local employer now and if it goes under, so does my town, which has the largest concentration of child poverty in the nation.
18.05.2025 14:16 — 👍 419 🔁 97 💬 4 📌 0
"I'm still in shock. I know that there have been political issues around Harvard in recent weeks, but antibiotic resistance isn't one of them." My conversation with Harvard microbiologist @baym.lol, one of many researchers there who just lost millions in fed. grants. www.wbur.org/news/2025/05...
16.05.2025 22:59 — 👍 428 🔁 243 💬 11 📌 19Our paper explaining why Gihawi et al. failed to prove an error in the normalization used by the 2020 cancer #microbiome analysis now out as a Matters Arising in @asm.org #mSystems (w/ @george-austin.bsky.social) 🖥️ 🧬
Thread explaining the key points below.
journals.asm.org/doi/10.1128/...
Our paper explaining why Gihawi et al. failed to prove an error in the normalization used by the 2020 cancer #microbiome analysis now out as a Matters Arising in @asm.org #mSystems (w/ @george-austin.bsky.social) 🖥️ 🧬
Thread explaining the key points below.
journals.asm.org/doi/10.1128/...
Come and work with me!
The Nature Micro team is expanding and we're looking for someone to champion microbial ecology, plant micro & related areas for the journal
Knowledge of microbial ecology/plant micro is desirable but we're open to applications from all microbiologists
Link below 👇
Congrats!
04.04.2025 20:19 — 👍 2 🔁 0 💬 0 📌 0