Amazing work by Takeo (@takeo-narita.bsky.social) on these comprehensive analysis and many new insights.
13.12.2024 13:07 — 👍 0 🔁 0 💬 0 📌 0@choudharylabcpr.bsky.social
@choudharylabcpr.bsky.social
Amazing work by Takeo (@takeo-narita.bsky.social) on these comprehensive analysis and many new insights.
13.12.2024 13:07 — 👍 0 🔁 0 💬 0 📌 0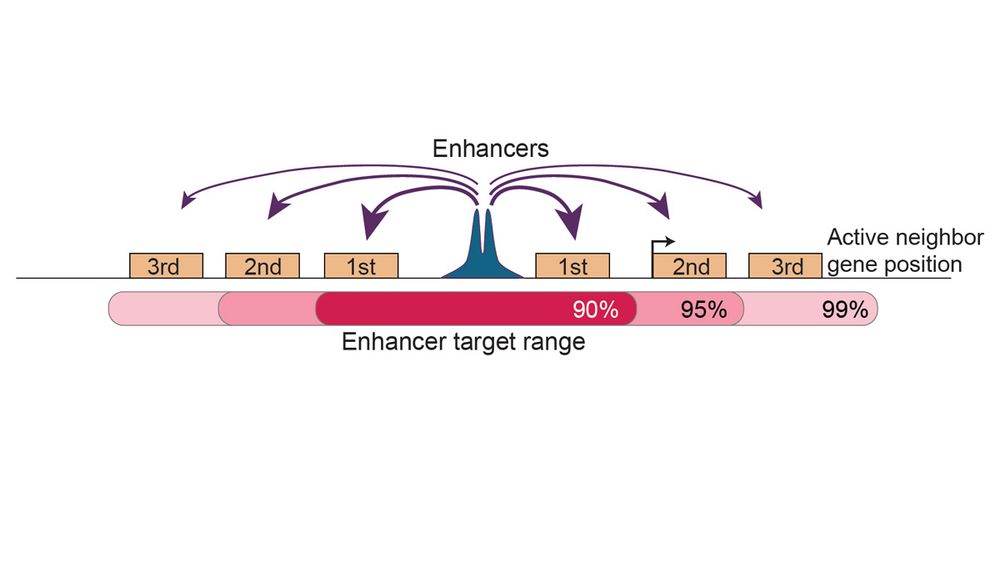
We propose that gene-skipping by enhancers are the exception, not the rule. Linear enhancer-promoter distance and coactivator requirement of genes can predict enhancer targets with accuracy rivalling state-of-the-art experimental approaches.
13.12.2024 13:07 — 👍 1 🔁 0 💬 1 📌 0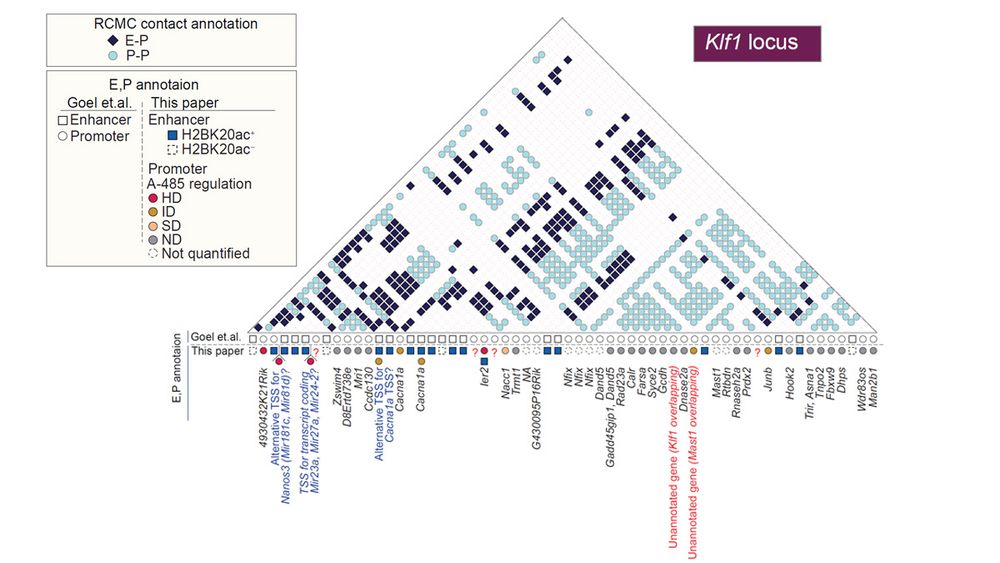
Multi-way enhancer-promoter interactions in ultra-deep chromatin contact mapping analyses don't indicate co-regulation of multiple promoters. Within multi-way interacting promoters, enhancers regulate proximal promoter but not distal ones.
13.12.2024 13:07 — 👍 0 🔁 0 💬 1 📌 0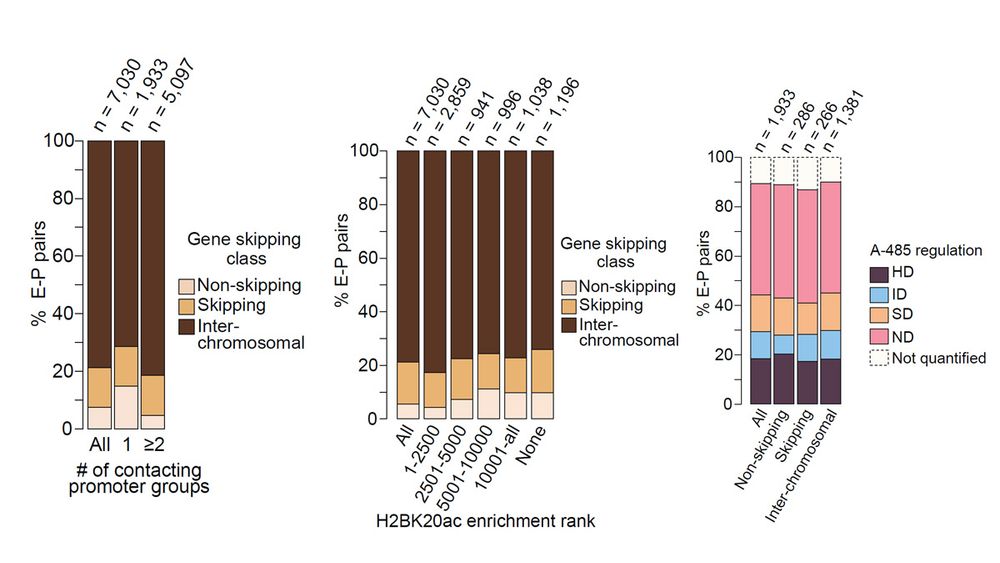
Attributes of enhancer targets identified by mapping eRNA-PROMPT RNA interactions by RIC-seq are fundamentally different from enhancer targets identified by CRISPRi.
13.12.2024 13:07 — 👍 0 🔁 0 💬 1 📌 0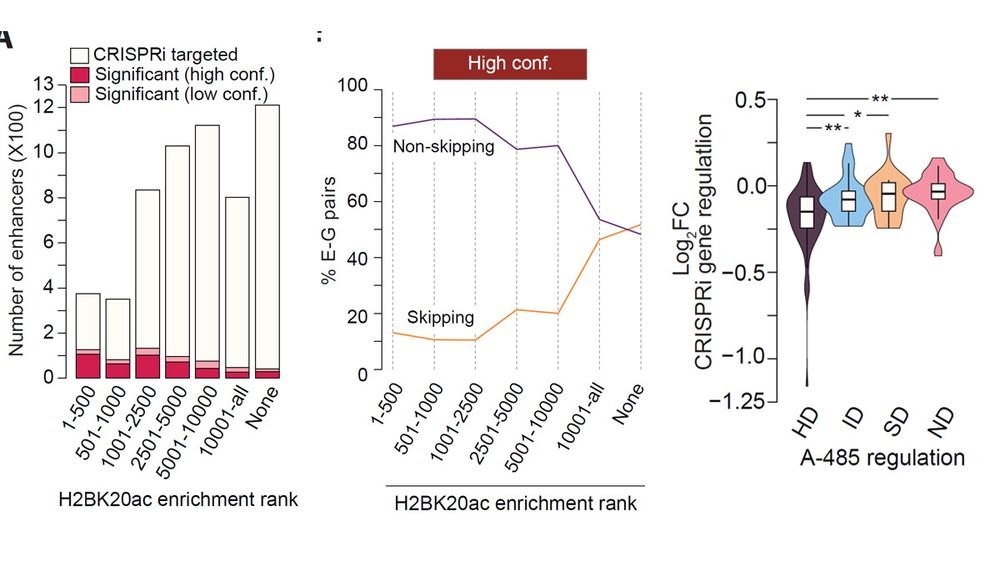
Gene-skipping and non-skipping enhancers differ in chromatin features, target gene distance, activation strength, and coactivator requirements. High H2BNTac strongly predict functional non-skipping enhancers, while skipping enhancers show weaker regulation and higher FDRs.
13.12.2024 13:07 — 👍 1 🔁 0 💬 1 📌 0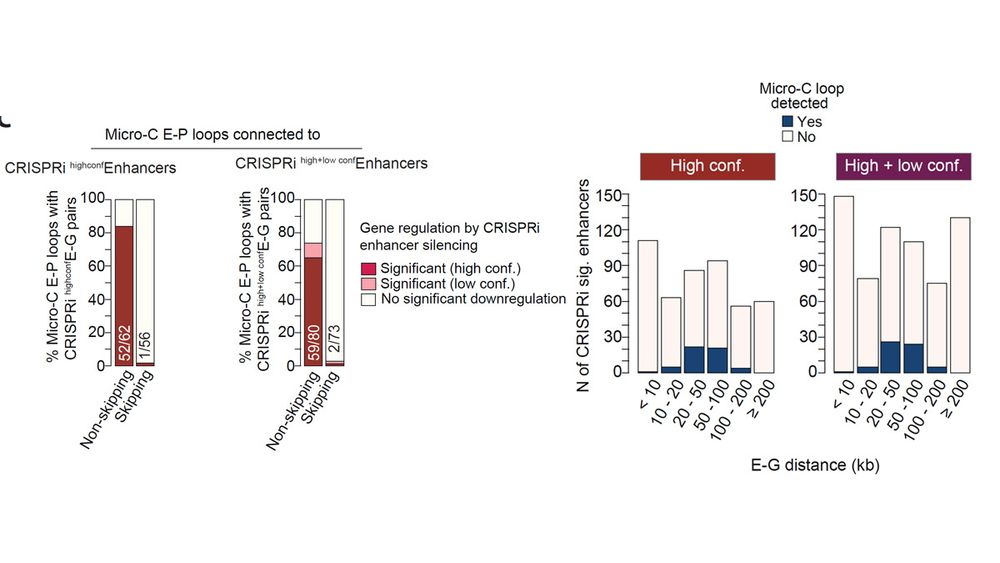
Gene-skipping enhancer-promoter contacts are rarely confirmed as being functional by CRISPRi screening. Conversely, CRISPRi identified gene-skipping enhancers also seldom show physical interactions.
13.12.2024 13:07 — 👍 0 🔁 0 💬 1 📌 0
Do enhancers often skip active genes? Our other recent preprint revises current models, that enhancers frequently and by design skip active genes to activate distal targets. Instead simple rules can predict enhancer and GWAS targets. www.biorxiv.org/content/10.1...
13.12.2024 13:07 — 👍 1 🔁 0 💬 1 📌 0Great effort by Takeo and the rest of the team in getting together this work.
09.12.2024 15:04 — 👍 0 🔁 0 💬 0 📌 0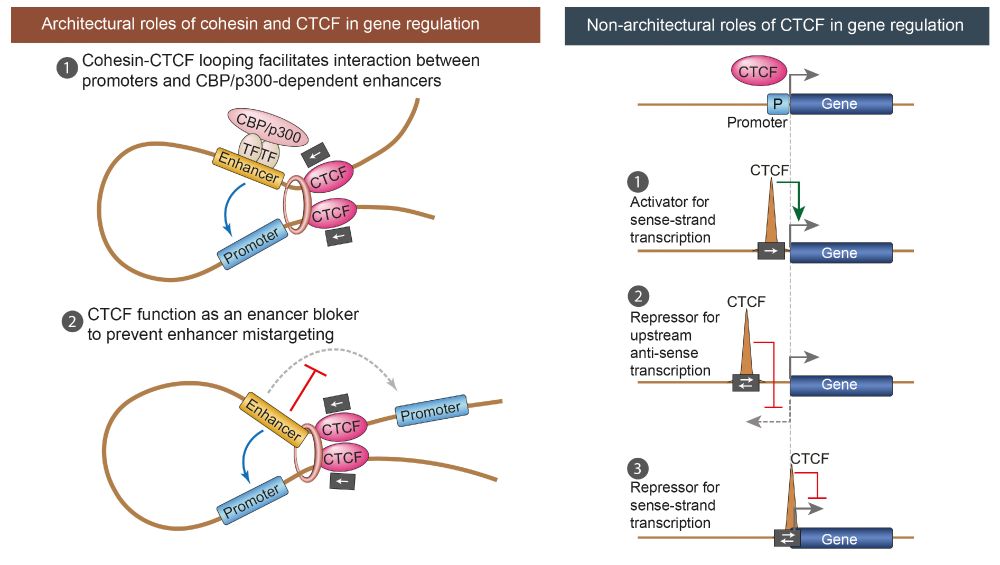
Our findings resolve several long-standing conundrums regarding the involvement of cohesin and CTCF in gene regulation, and we present a unified model for their combined and individual roles acting directly near promoters or in concert with p300/CBP-dependent enhancers.
09.12.2024 15:04 — 👍 1 🔁 0 💬 1 📌 0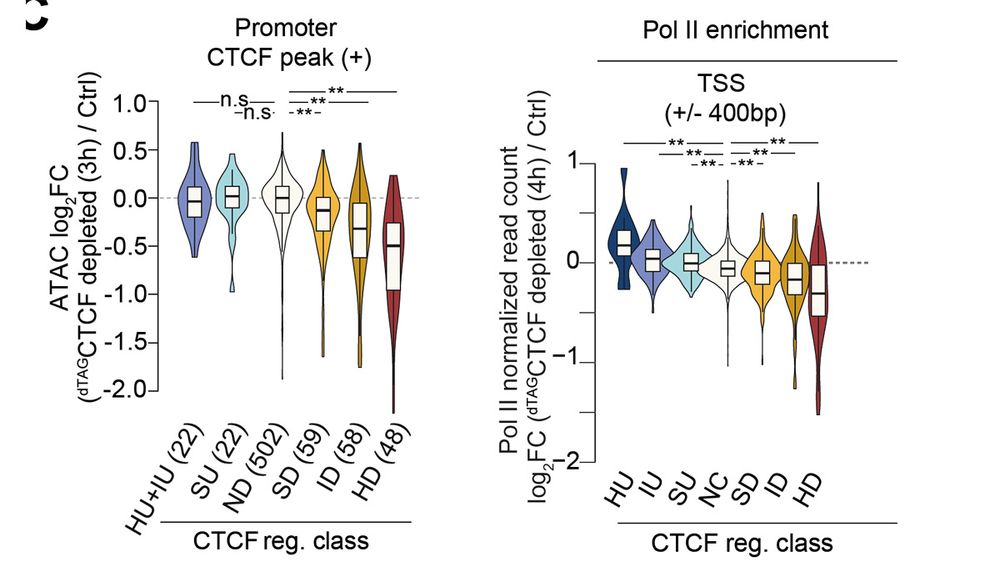
For the genes regulated by CTCF at the promoter, CTCF directly impacts DNA accessibility and downstream recruitment of PolII.
09.12.2024 15:04 — 👍 0 🔁 0 💬 1 📌 0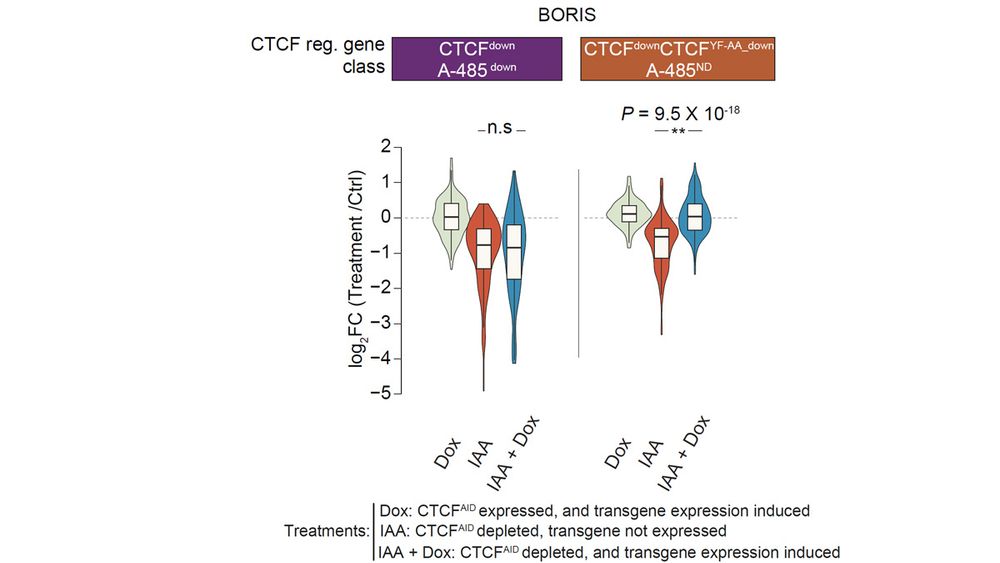
The CTCF-like protein BORIS is also unable to anchor cohesin. Re-analysis of published data with this new perspective show, that unlike CTCF, BORIS has no impact on enhancer-regulated genes. In contrast, BORIS can selectively rescue expression of promoter-activated genes.
09.12.2024 15:04 — 👍 0 🔁 0 💬 1 📌 0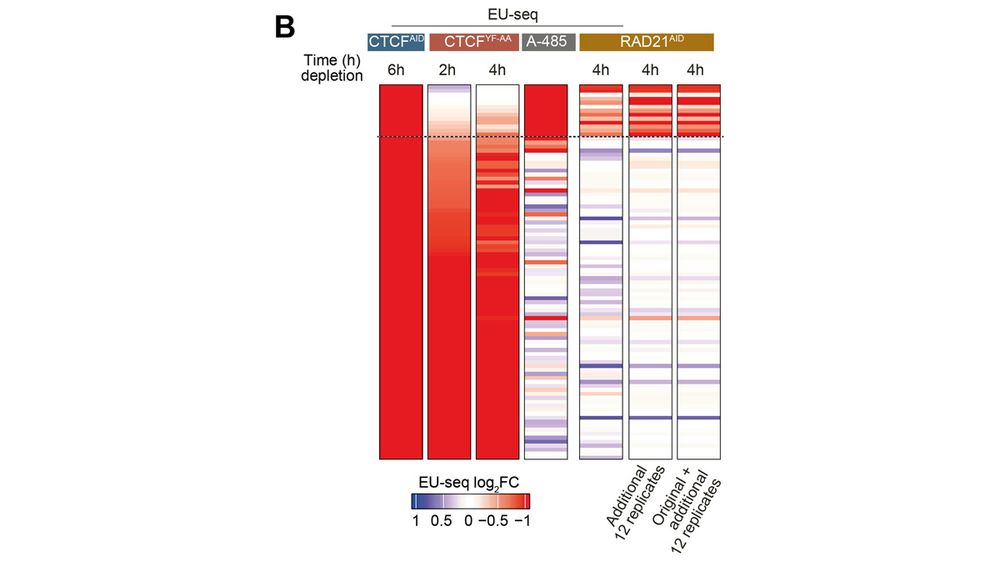
Using a cohesin-loop-deficient CTCF mutant, we reveal that CTCF is a position- and orientation-dependent transcription activator at promoters. It directly activates hundreds of essential housekeeping genes—independently of cohesin and enhancers—even in plasmid reporter assays.
09.12.2024 15:04 — 👍 0 🔁 0 💬 1 📌 0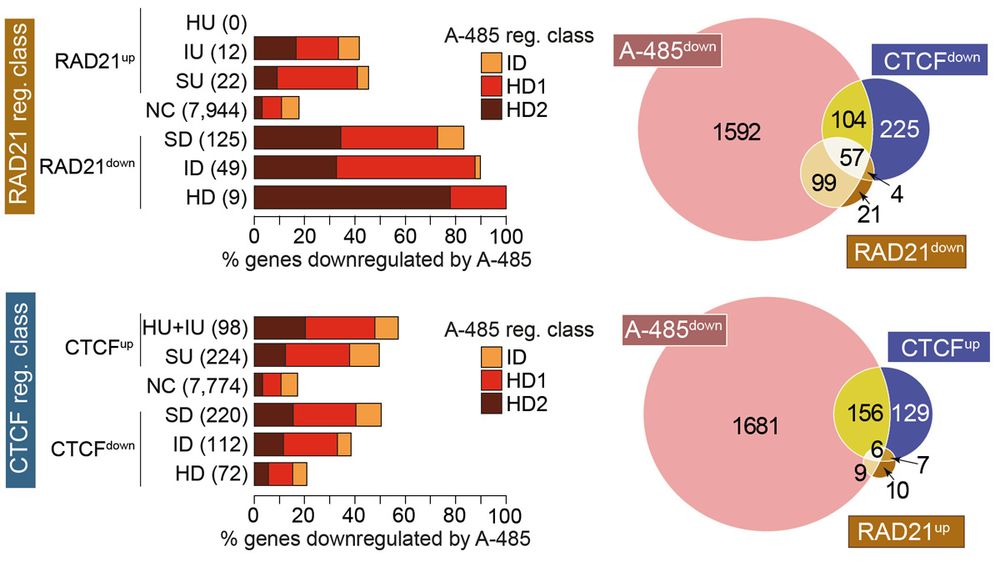
Among diverse types of posited enhancers, cohesin seems to uniquely facilitate enhancer-mediated gene regulation through p300/CBP-dependent enhancers.
09.12.2024 15:04 — 👍 0 🔁 0 💬 1 📌 0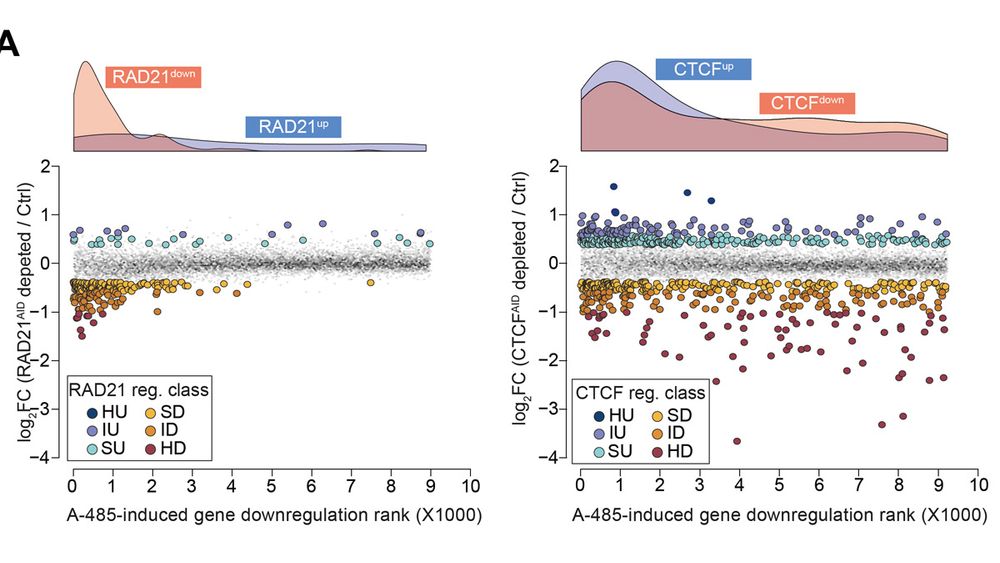
Acute depletion of cohesin and CTCF dysregulate several hundreds of genes – far more than previously appreciated. Many of these are subtle, but as confirmed through several repetitions, they are consistent.
09.12.2024 15:04 — 👍 0 🔁 0 💬 1 📌 0
How do cohesin and CTCF control gene expression and what are their links to enhancers? Our new preprint unravels their shared and unique roles: from precise enhancer targeting to direct transcriptional activator functions of CTCF!
#Genomics #CTCF #Cohesin
www.biorxiv.org/content/10.1...