It was a great pleasure to know and interact with Amos, he will be greatly missed.
02.12.2025 09:02 — 👍 2 🔁 0 💬 0 📌 0It was a great pleasure to know and interact with Amos, he will be greatly missed.
02.12.2025 09:02 — 👍 2 🔁 0 💬 0 📌 0
CATH turns 30 years old this year!
We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

www.nature.com/articles/s41... From remote matches to @rfamdb.bsky.social models to the discovery of novel *vertebrate* riboswitches!
19.08.2025 01:20 — 👍 5 🔁 3 💬 1 📌 0
Beyond homology: This mind blowing work demonstrates the power of genomics driven analogy for discovery in the era of structure predictions.
The diversity of defense systems emerged thx to "guilt by association approaches". New analogies to uncover molecular novelties 👇.
👏 Nitzan, Rotem & al.

New impact case study outlines the impressive economic & scientific value of UniProt - the world-leading open data resource for protein sequence & functional information.
Users save up to 219 hours per year, equivalent to net benefits of up to €5,475 per user.
www.ebi.ac.uk/about/news/a...

Pfam 37.4 318 new families 4 new clans

Content Over 24,736 entries describing protein families, domains and repeats.
🚀 HUGE NEWS: Pfam 37.4 is LIVE!
318 brand-new protein families + 4 new clans! 🧬
Total collection now: 24,736 entries! 📈
Get exploring via InterPro website, API & Pfam FTP!
💻🧪
We would welcome applicants who wish to advance @pfamdb.bsky.social and/or @interprodb.bsky.social
21.05.2025 08:53 — 👍 2 🔁 0 💬 0 📌 0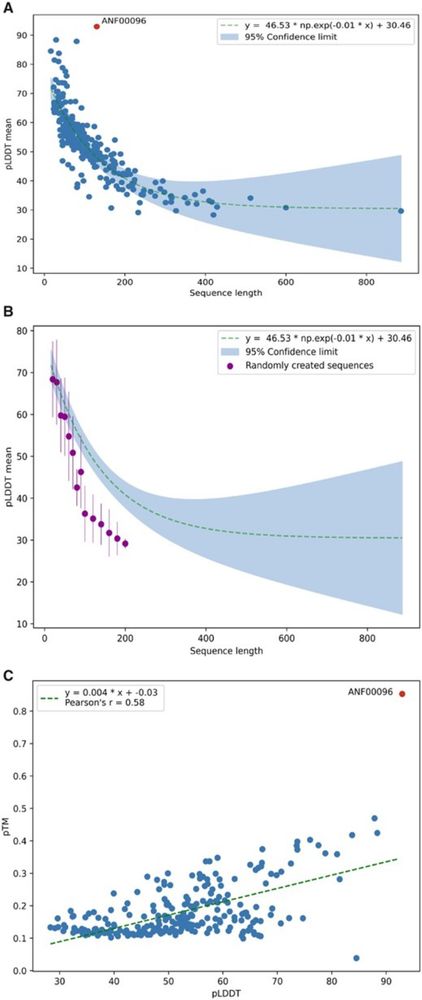
Surprise! It folds into an Alpha-helix. Hmm, short peptides get unreasonably high pLDDT scores with AlphaFold. We see mainly alpha helix predictions for short peptides of random sequences or spurious mistranslated protein sequences: academic.oup.com/bioinformati...
Still looks like a great paper.
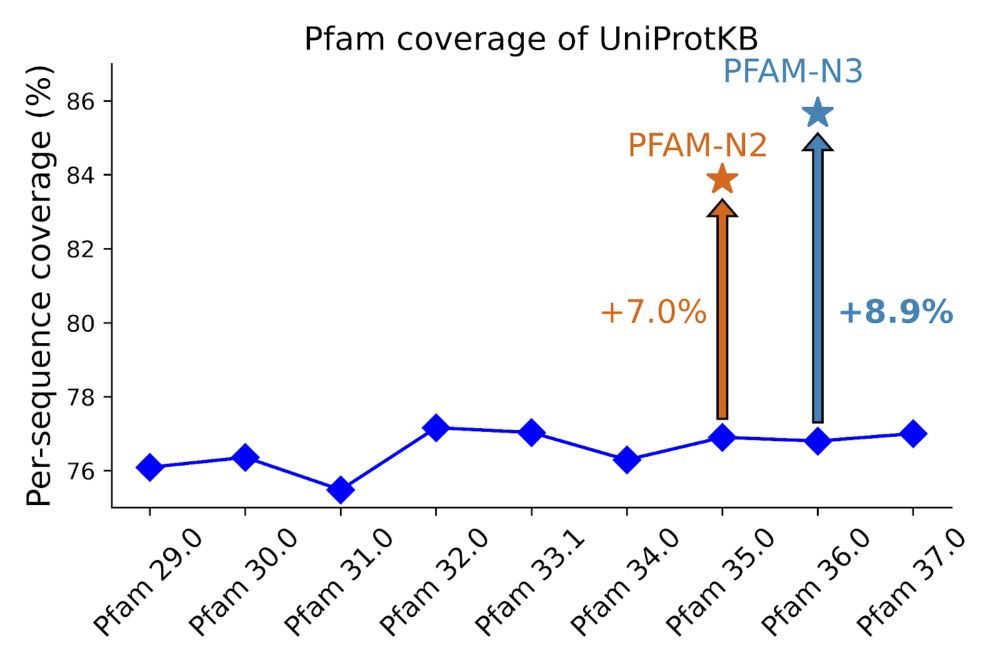
Hi, InterPro-N is casically Pfam-N but trained across all of InterPro. So Pfam-N would be taking the InterPro-N subset that matched Pfam. Some explanation here: xfam.wordpress.com/2024/05/31/p...
29.04.2025 08:32 — 👍 2 🔁 0 💬 0 📌 0I'm really excited about this InterPro release. Now including BFVD virus structures, InterPro-N matches and TED domains.
24.04.2025 20:38 — 👍 0 🔁 0 💬 1 📌 0
🔎 Isopeptor is available as a #Python-based tool that can be accessed via the command line, a Python API, or Google Colaboratory. Use it for automated isopeptide bond detection & validation in protein structures!
Google Colab: https://t.ly/D4Lny
GitHub:
"Isopeptor" introduces a #machinelearning-based approach to systematically identify isopeptide bonds in protein models. By integrating structure-guided template matching with a logistic regression classifier, Isopeptor enhances structural annotation and model accuracy.
24.03.2025 12:02 — 👍 1 🔁 1 💬 1 📌 0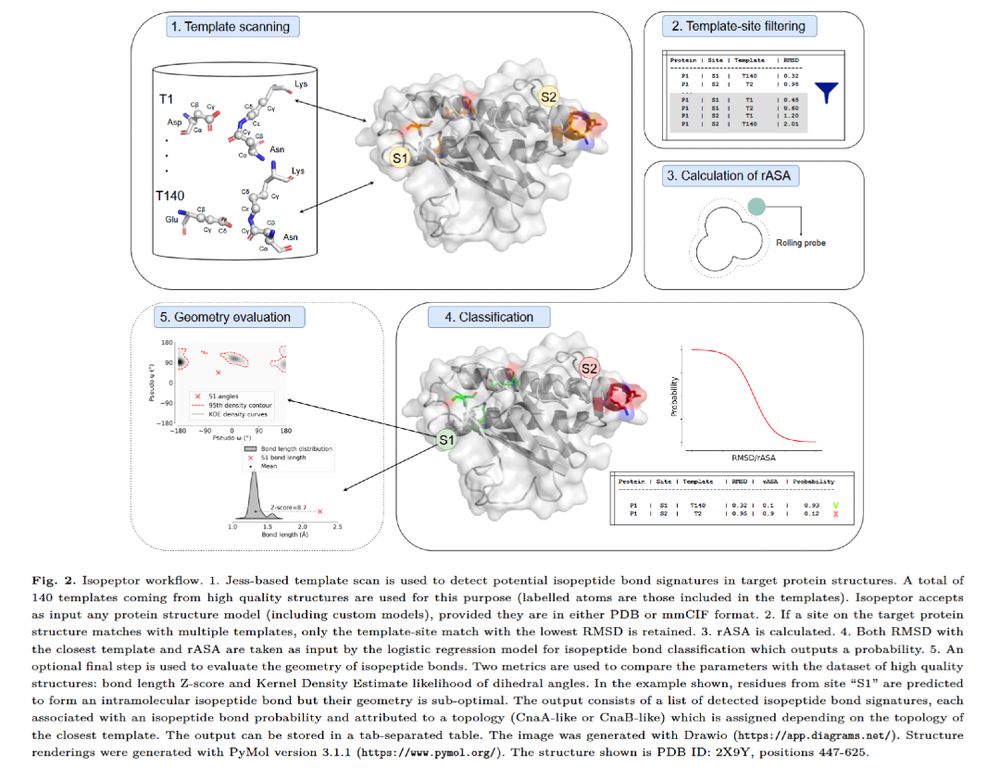
🧩A new article in Bioinformatics Advances: "Isopeptor: A tool for detecting intramolecular isopeptide bonds in protein structures"
Read more here: https://doi.org/10.1093/bioadv/vbaf049

📢 We’re looking for a new Team Leader!
@enasequence.bsky.social is recruiting a Team Leader to shape the strategy and direction for the ENA and Biosamples open data resources.
Interested?👇
Well this new paper is a real pleasure! Not every Masters student gets to publish their work, so special congratulations to Olivia Pratt.
www.csbj.org/article/S200...
She shows that AlphaFold 2, but not other tools, has a real blind spot with perfect sequence repeats.
(1/n)

Photo of Cambridge River and view over King's College with a group of people punting.

Photo of the Heart and Lungs Research Institute on Cambridge Biomedical Campus. The building is bronze-colour and in front of a pond.
🚨🌱 Interested in making scientific computing greener? Here is a chance to do just that in Cambridge: 2 open research roles to work with me and the Green Algorithms project.
More details about the roles below👇 and at www.lannelongue-group.org/join/
⏳ Closing: Feb 4 for one, Feb 17 for the other
Amazing - the day Steve Jobs and Tim Berners Lee ‘met’. open.substack.com/pub/stephenf...
17.01.2025 14:47 — 👍 4 🔁 3 💬 0 📌 0Looks like a really useful development!
17.01.2025 13:03 — 👍 4 🔁 0 💬 0 📌 0As a Claude user I found the system prompt a very interesting read.
02.01.2025 09:18 — 👍 0 🔁 0 💬 0 📌 0
You mean this photo?
01.01.2025 18:01 — 👍 5 🔁 1 💬 1 📌 0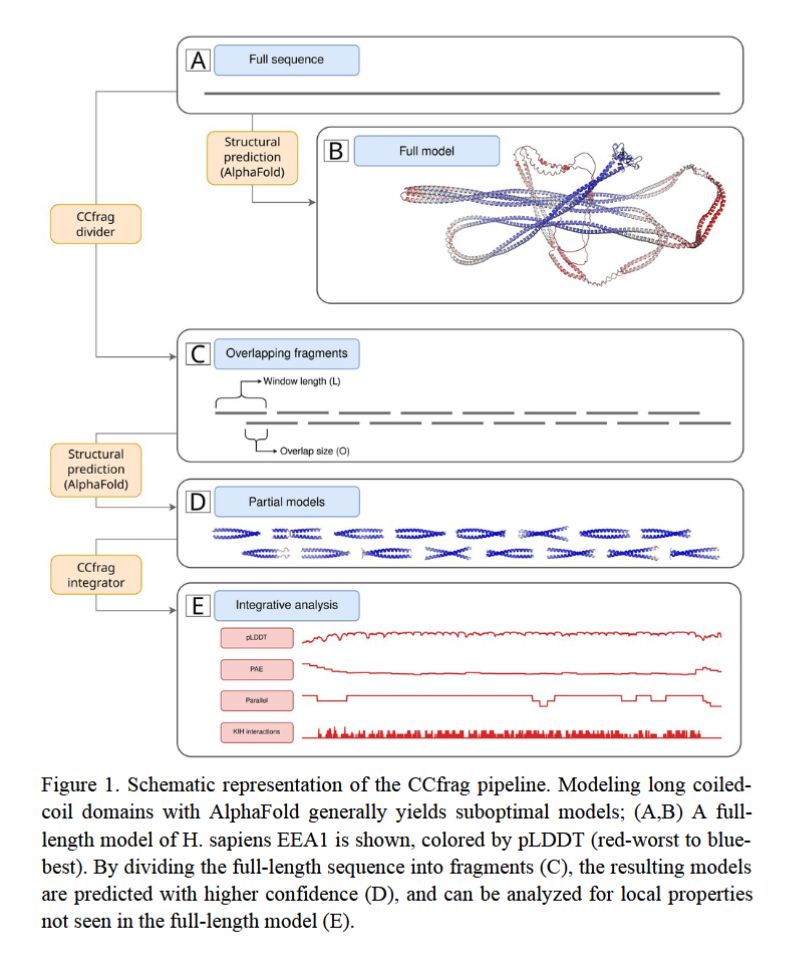
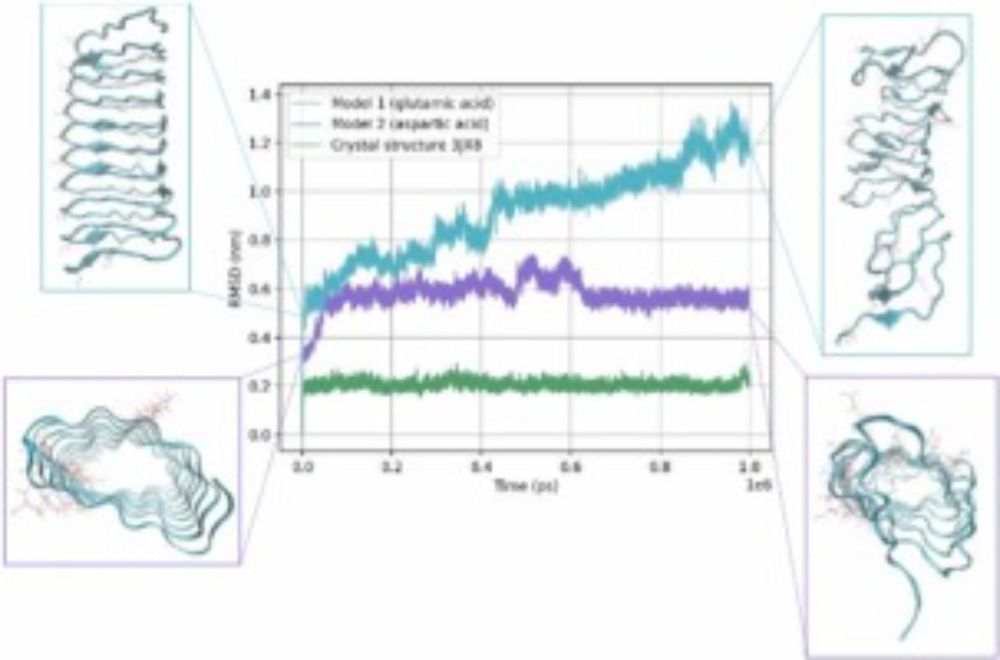
🧬 "CCfrag: Scanning folding potential of coiled-coil fragments with AlphaFold" introduces a fragment-based approach to improve protein structure prediction by leveraging overlapping sequence windows for better modeling of coiled-coils.
Read more: doi.org/10.1093/bioa... #ProteinFolding #AlphaFold
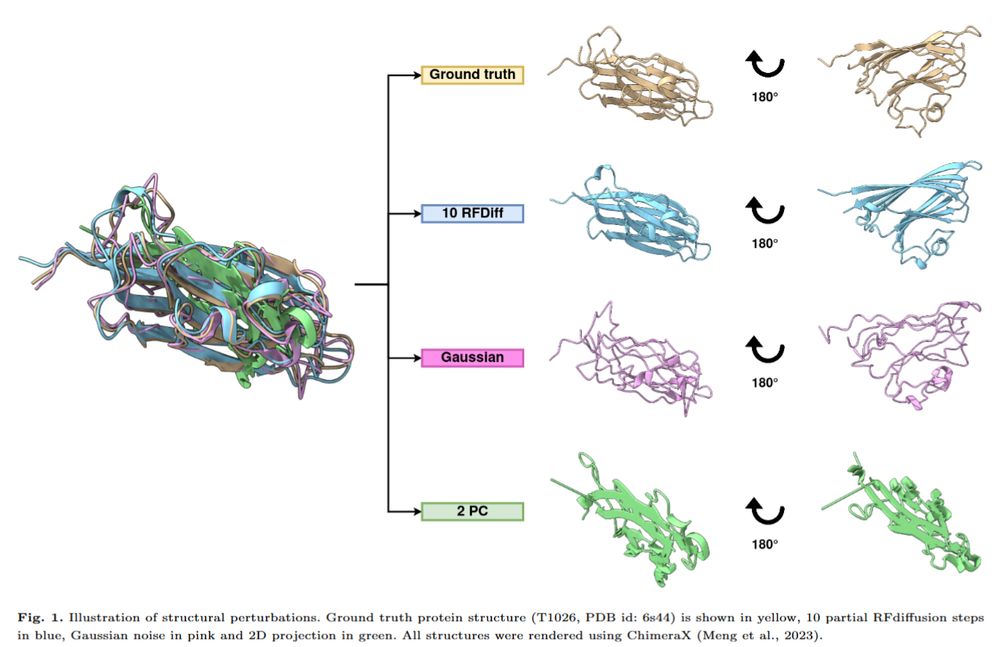
🔬 How does #AlphaFold2 perform with minimal sequence input?
"Dissecting AlphaFold2’s capabilities with limited sequence information" reveals insights into its ability to predict #ProteinStructures using structural templates without deep MSAs.
Read more: doi.org/10.1093/bioa...
New work from the group showing that you can rescue poor quality alphafold models using AF models of homologous proteins
19.12.2024 15:37 — 👍 1 🔁 0 💬 0 📌 0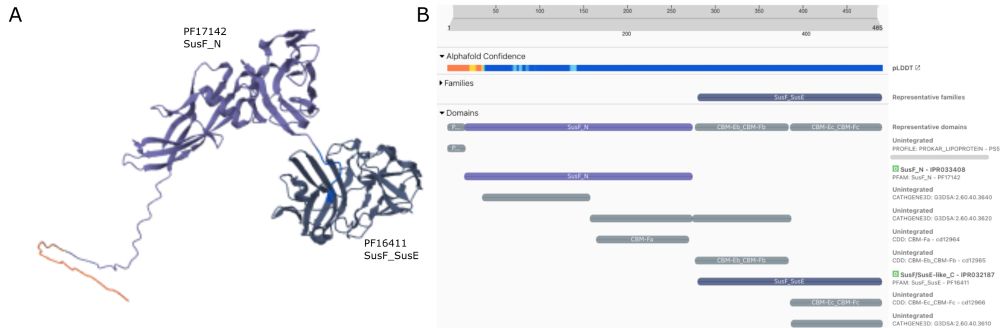
🧬 What's new in Pfam? Beta propellers uncovered through smart deduplication, fresh perspectives on haloarchaeal proteins, and more! Dive into our latest blog: xfam.wordpress.com/2024/12/09/p...
09.12.2024 09:58 — 👍 8 🔁 2 💬 0 📌 0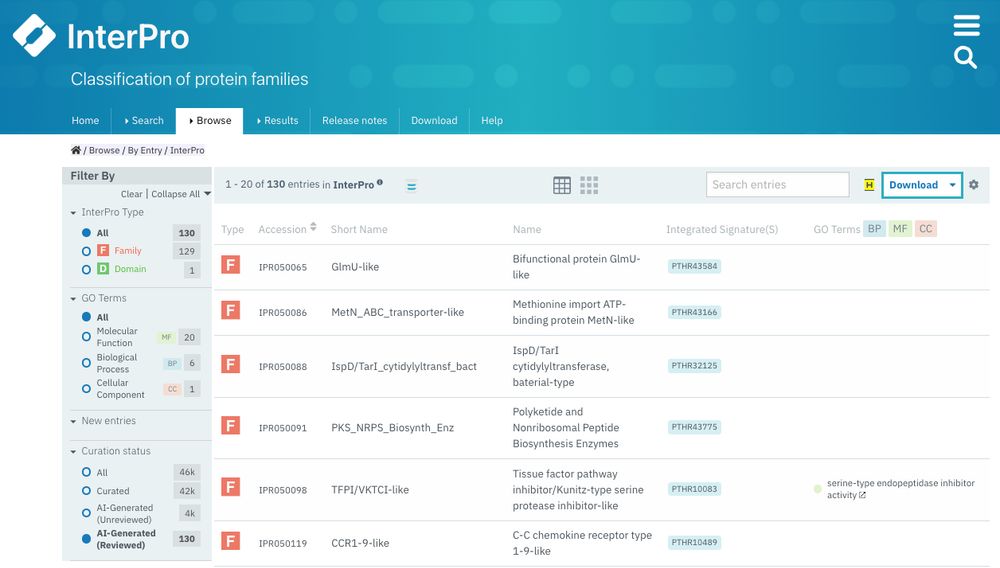
AI-filter in InterPro
📝 Just published: Our latest blog walks you through InterPro's major updates - from redesigned protein views to improved data exploration. Read more: proteinswebteam.github.io/interpro-blo...
09.12.2024 10:05 — 👍 5 🔁 1 💬 0 📌 0A nice study by @atsocf.bsky.social in my lab who shows how to rescue poor quality AlphaFold models using within family templates
09.12.2024 14:32 — 👍 1 🔁 0 💬 0 📌 0Very nice work. We have just added a new @pfamdb.bsky.social entry for the Pyrenoid shell proteins DiatomPyrShell PF25192. Will be available in a future Pfam release.
30.11.2024 08:10 — 👍 1 🔁 0 💬 1 📌 0I definitely need this in my stream :-)
23.11.2024 11:42 — 👍 1 🔁 0 💬 0 📌 0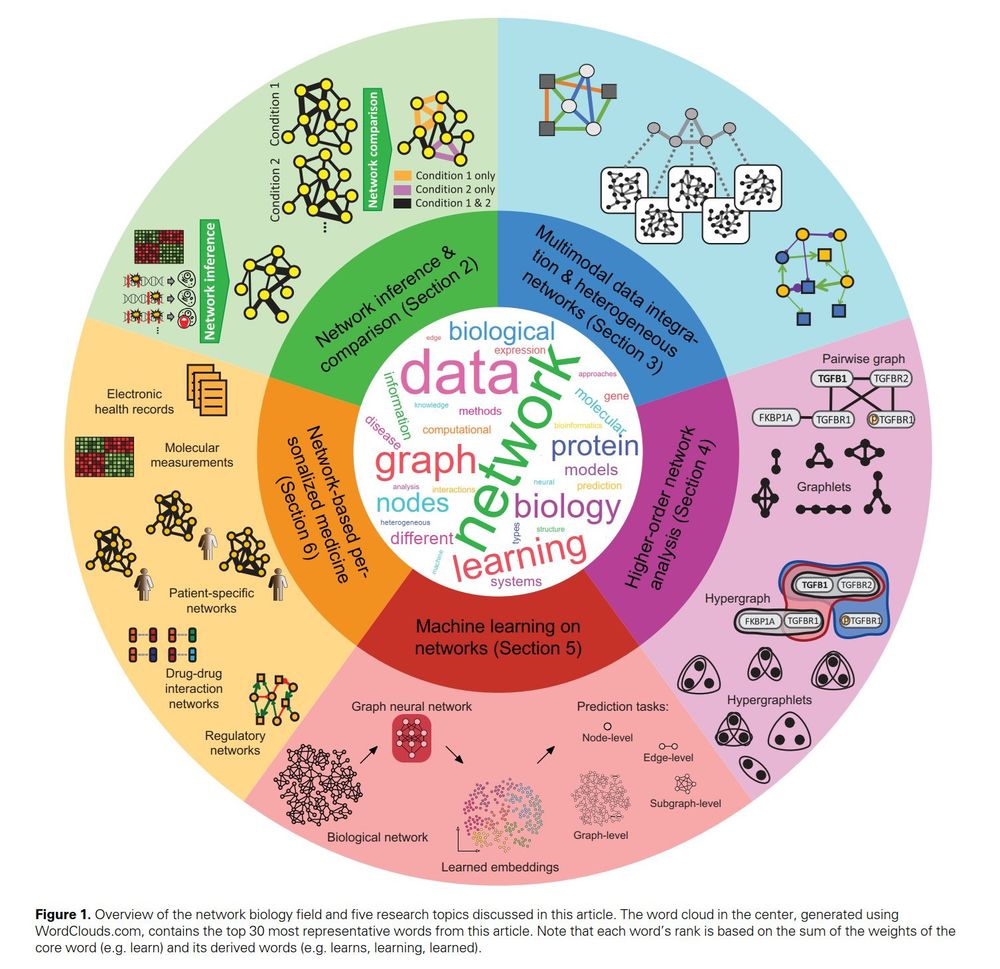
ICYMI, August 2024: "Current and future directions in network biology" shares insights into advancing cellular function and disease research through #MachineLearning, multimodal data integration, and personalized medicine.
Read it here: doi.org/10.1093/bioa...
#NetworkBiology #SystemsBiology
I will start my Bluesky life by sharing an opportunity for those who are interested in researching fungi and how they adapt to their environment. Awards of up to £3 million available, more details at the link below or in our webinar on 27th November.
wellcome.org/grant-fundin...