Congratulations, Matthieu! 🥳
18.06.2025 16:04 — 👍 1 🔁 0 💬 0 📌 0
Very happy to release AutoMartiniM3 for Martini3 to automatize CG modelling of small molecules and fragments.
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social
11.06.2025 06:49 — 👍 37 🔁 11 💬 1 📌 2

Martini workshop registration now open !!!!
See cgmartini.nl for details and how to apply.
Looking forward to seeing you in Groningen, Aug 11-15th.
27.05.2025 07:55 — 👍 12 🔁 8 💬 0 📌 4

Pre-announcement: MARTINI tutorial workshop, August 11-15th 2025 in Groningen, The Netherlands !!
Learn basic and advanced Martini from the cocktail masters themselves. Registration will open soon.
28.04.2025 09:06 — 👍 23 🔁 12 💬 0 📌 0
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
11.04.2025 08:35 — 👍 358 🔁 142 💬 12 📌 20
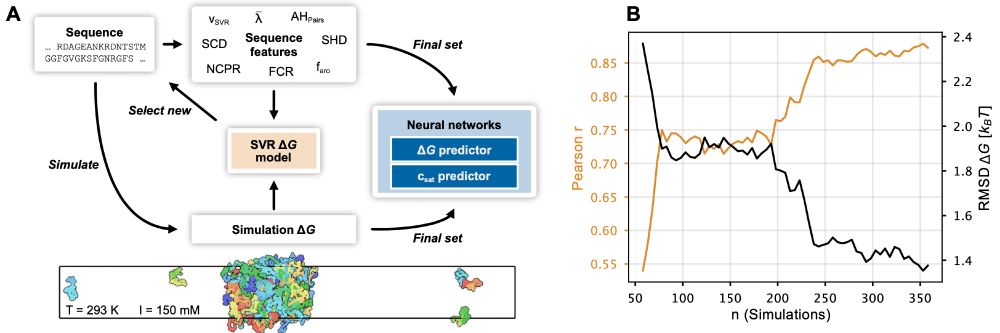
Our paper on prediction of phase-separation propensities of disordered proteins from sequence is now published:
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
25.03.2025 17:55 — 👍 71 🔁 17 💬 1 📌 1
Careers
Careers
We’re looking for fellow nerds to help us make disordered proteins druggable. Love #IDPs, #MD, #NMR, #AI, #DrugDiscovery, or #MassSpec? Let’s talk.
@bindresearch.org is hiring across the board: bindresearch.org/careers/
#compchem #compbio #NMRChat #TeamMassSpec #chemjobs @chemjobber.bsky.social
24.03.2025 10:51 — 👍 26 🔁 16 💬 0 📌 0
We are organizing a conference for young researchers at the intersection between physics & biology: www.embl.org/about/info/c... @embl.org @events.embl.org @intcha.bsky.social
Please consider applying/registering -- the deadline is already in one week!
24.03.2025 14:35 — 👍 44 🔁 27 💬 0 📌 0

Airplane
Traveling to San Diego for ACS. Really looking forward to the meeting ☺️.l!
22.03.2025 14:25 — 👍 9 🔁 0 💬 1 📌 0
CALVADOS now has parameters for phosphorylated amino acids
@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...
21.03.2025 07:01 — 👍 50 🔁 16 💬 1 📌 2
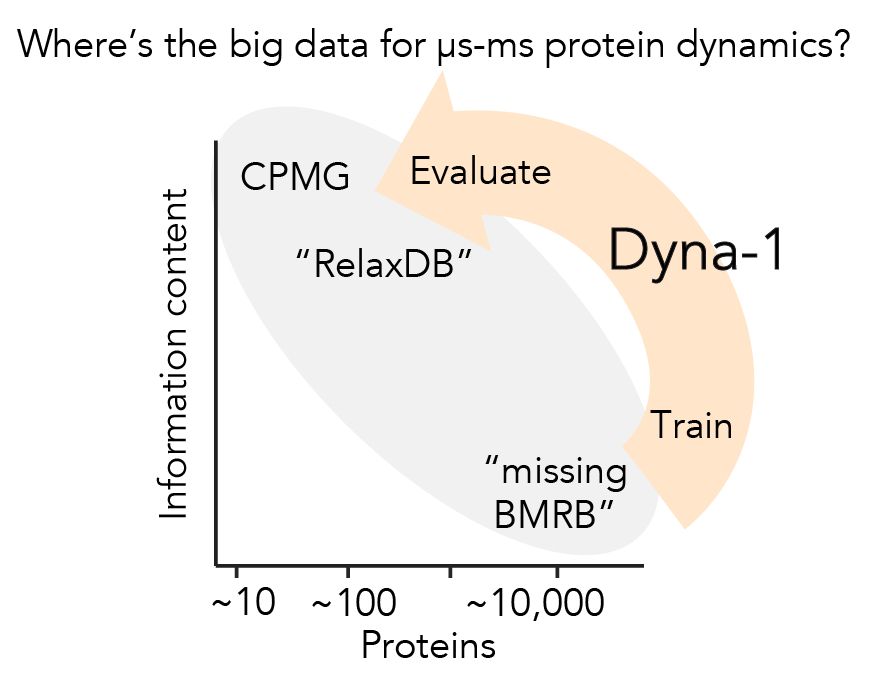
Protein dynamics was the first research to enchant me >10yrs ago, but I left in PhD bc I couldn't find big experimental data to evaluate models.
Today w @ginaelnesr.bsky.social, I'm thrilled to share the big dynamics data I've been dreaming of, and the mdl we trained w them: Dyna-1.
📝: rb.gy/de5axp
20.03.2025 15:02 — 👍 85 🔁 25 💬 2 📌 1
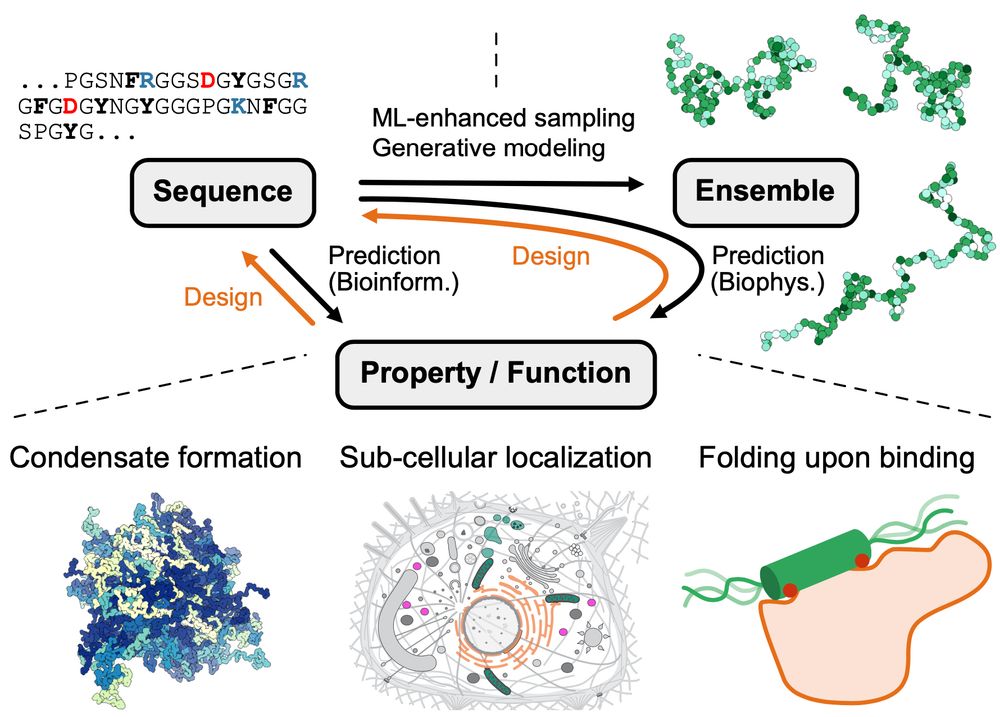
Figure from the paper illustrating sequence–ensemble–function relationships for disordered proteins. ML prediction (black) and design (orange) approaches are highlighted on the connecting arrows. Prediction of properties/functions from sequence (or vice versa, design) can include biophysics approaches via structural ensembles, or bioinformatics approaches via other hetero- geneous sources. The lower panels show examples of properties and functions of IDRs for predictions or design targets. ML, machine learning; IDRs, intrinsically disordered proteins and regions.
Our review on machine learning methods to study sequence–ensemble–function relationships in disordered proteins is now out in COSB
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
12.03.2025 21:37 — 👍 90 🔁 27 💬 0 📌 1
Go and work with @reidalderson.bsky.social !!!
11.03.2025 07:33 — 👍 2 🔁 0 💬 1 📌 0
New Preprint ‼️ Work led by the incredible @ananyac2000.bsky.social 💃🏽💃🏽 Mpipi-T is finally here!!! 🥳🥳 👇🏽👇🏽
www.biorxiv.org/content/10.1...
06.03.2025 22:10 — 👍 25 🔁 9 💬 0 📌 0
CALVADOS 🤝 PEG
Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs
06.03.2025 22:03 — 👍 41 🔁 7 💬 2 📌 2
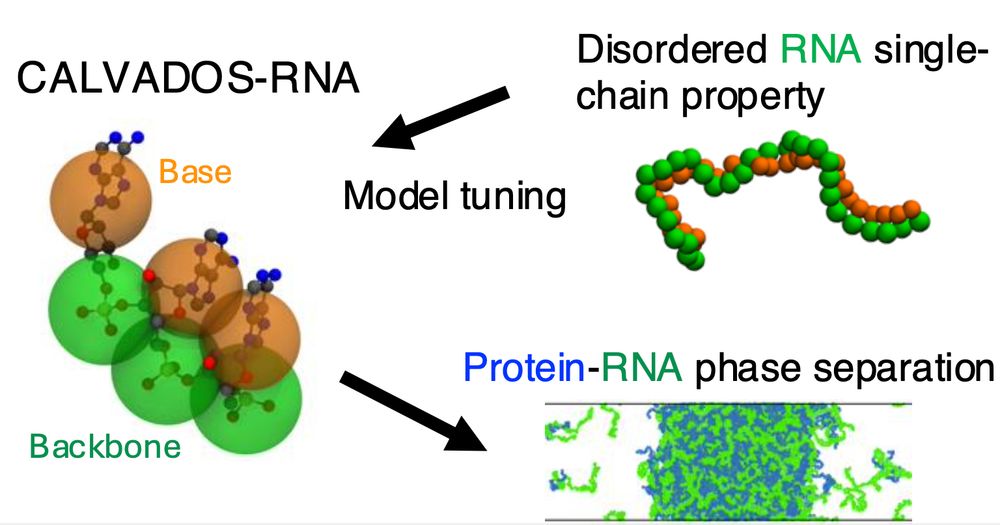
Table of Contents figure showing the CALVADOS-RNA model and a snapshot from a mixed protein-RNA condensate
CALVADOS-RNA is now published
doi.org/10.1021/acs....
This is a simple model for flexible RNA that complements and works with the CALVADOS protein model. Work led by Ikki Yasuda who visited us from Keio University.
Try it yourself using our latest code for CALVADOS
github.com/KULL-Centre/...
26.02.2025 19:11 — 👍 67 🔁 20 💬 1 📌 0
Congratulations!!!
24.02.2025 16:44 — 👍 2 🔁 0 💬 0 📌 0

DPhil in Biomedical and Clinical Sciences (Oxford-GSK) | University
About the courseThe DPhil in Biomedical and Clinical Sciences (Oxford-GSK) aims to address an unmet need among clinicians for training in data science and translational medicine. The course provides
Great opportunity for clinicians to gain early-career experience in a fully-funded, translational medicine programme in Oxford.
If you know anyone interested, please share this link. There is a wide range of projects, including mine. Deadline Fr 14 March 2025
www.ox.ac.uk/admissions/g...
21.02.2025 16:58 — 👍 2 🔁 4 💬 0 📌 1
The BioEmu-1 model and inference code are now public under MIT license!!!
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
19.02.2025 20:17 — 👍 103 🔁 39 💬 2 📌 2
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
PhD student in biophysics/biochemistry working on the interaction between the phosphorylated Tau protein and microtubules. Molecular dynamist. Tamer of IDRs and IDPs since 2022
(They/Them)
https://scholar.google.com/citations?user=4S1QUPgAAAAJ&hl=fr
Statistical mechanics & biophysics theorist. Emergent properties in biomolecules. Systems biology curious. Father, former athlete. Kansas State University Physics. Occasional appearance of Legos.
Biologist by training, basketball player at heart
Professor for Translational Epigenetics & Genome Architecture, University Medical Center Goettingen, Germany.
www.papantonislab.eu
PhD student at CBS Montpellier 🇫🇷
MD simulations of proteins, RNA and condensates 🧬💻
PhD student at Center for Brain and Disease, VIB, KU Leuven. Particularly interested in Neuroscience, Alzheimer's Disease, and Phase Separation.
Farmer | PhD Student at https://sites.google.com/view/boselab, IIT Kharagpur | RNA Binding Protein | Biomolecular Condensates | M.Tech at IIT Kharagpur | #Firstgen
At KITP on the UC Santa Barbara campus, researchers in theoretical physics and allied fields collaborate on questions at the leading edges of science.
www.kitp.ucsb.edu
Single Molecule Biophysics
Quantum Photonics
Postdoc @ ETH Zurich / SIB Swiss Institute of Bioinformatics | Protein bioinformatics
Professor in bioinformatics, Stockholm University. Protein structure lover ( interactions predictions, evolution ..). Using machine learning as a part of AI for Sciences for halv my life. In addition to succén, I sometimes rants about sailing or skiing.
Cooperation of @hitsters.bsky.social, @uniheidelberg.bsky.social & @kit.edu on bridging scales from molecules to molecular materials by multiscale simulation & machinelearning
Cell biology lab at Goethe University Frankfurt. Exploring subcellular architecture with super-resolution fluorescence microscopy. Focused on how organelles coordinate lipid and energy metabolism.
www.stephan-lab.de
(He/they) Physicist. ML scientist, PhD in AI for science. Barcelona. 🏳️🌈🏳️⚧️
Computational biophysicist @ Max Planck Institute for Polymer Research, Mainz.